-
Notifications
You must be signed in to change notification settings - Fork 16
3D_Nuclei_Clustering_Tool
Analyze the clustering behavior of nuclei in 3D images. The centers of the nuclei are detected. The nuclei are filtered by the presence of a signal in a different channel. They clustered with the density based algorithm DBSCAN. The nearest neighbor distances between all nuclei and those outside and inside of the clusters are calculated.
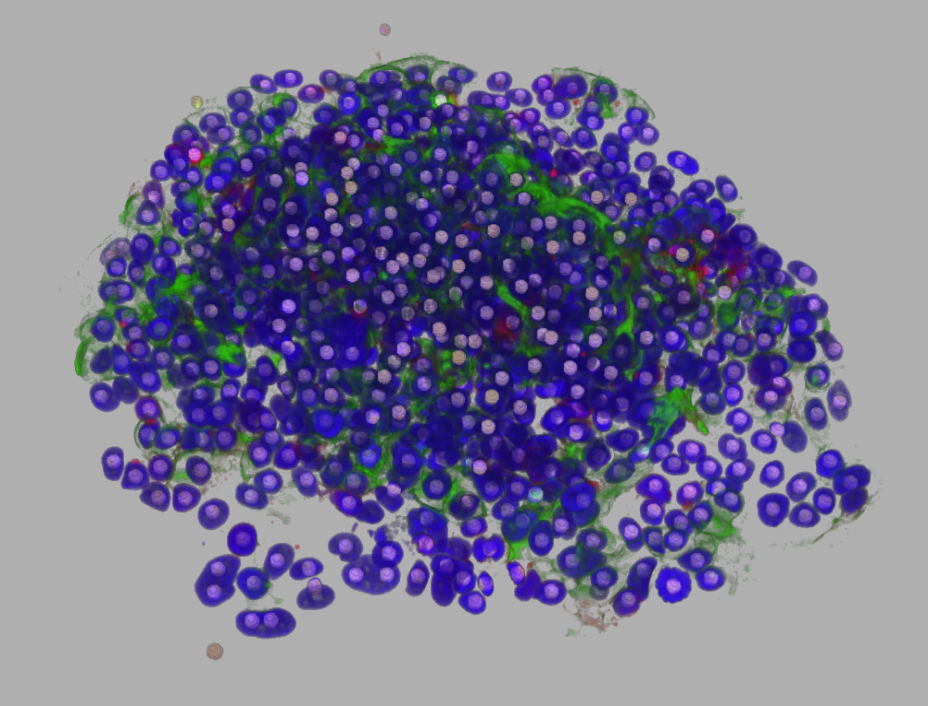
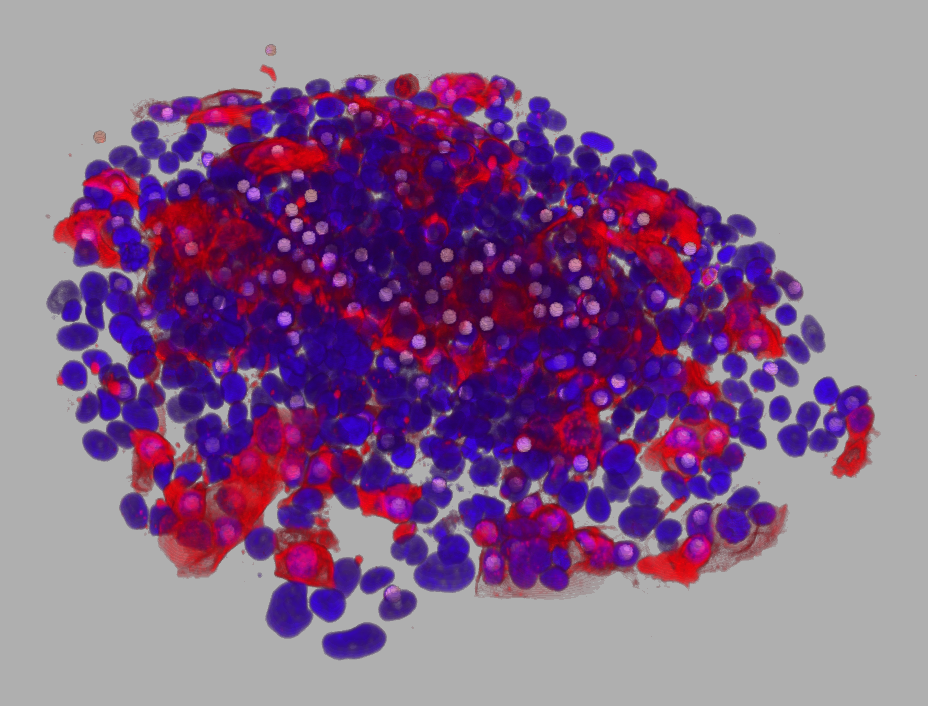


You can download an example data-set:
The source code in git-hub can be found here.
You must have the 3D ImageJ Suite [1] and FeatureJ installed. They are available via the FIJI-update-sites 3D ImageJ Suite and ImageScience.
To install the tool save the three files 3D_nuclei_clustering.ijm, dbscan_clustering_3D.py and nearest_neighbor_distances_3D.py into the folder macros/toolsets of your FIJI installation.
Select the "3D_nuclei_clustering" toolset from the >> button of the ImageJ launcher.
- the first button (the one with the image) opens this help page
- the
p-button runs the complete analysis on the active image - the
b-button runs the batch analysis on all images in a folder
The remaining buttons execute one of the steps of the analysis at a time. This can be helpful for finding the best option-values.
- the
d-button detects the nuclei in the image - the
f-button filters the nuclei, so that only nuclei with an intensity above a threshold in another channel remain. - the
c-button runs the cluster analysis. - the
n-button calculates the nearest-neighbor distances for the active table - the
v-button visualizes the nearest-neighbor connections - the images menu-button allows to download an example dataset and to open the contained images
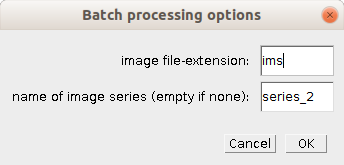
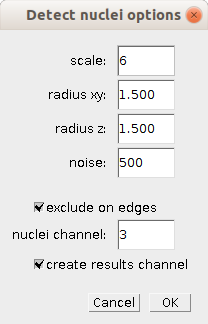
Right-click one of the buttons b, d, f or c to open the options-dialog of each analysis step.
- image file-extension
- The file extension of the input images
- name of the image series (empty if n none)
- The series in the image file that will be used. If your image files do not contain series enter an empty text.
- scale
- The smoothing scale of the [LoG filter](https://imagescience.org/meijering/software/featurej/laplacian/) used in the detection of the nuclei.
- radius xy
- The radius of the spots in the x and y dimension for the [3D maximum detection](https://imagejdocu.tudor.lu/tutorial/plugins/3d_maxima_finder).
- radius z
- The radius of the spots in the z dimension for the [3D maximum detection](https://imagejdocu.tudor.lu/tutorial/plugins/3d_maxima_finder).
- noise
- The noise value for the [3D maximum detection](https://imagejdocu.tudor.lu/tutorial/plugins/3d_maxima_finder). Only local maxima above the noise-value are detected.
- exclude on edges
- If selected no nuclei will be detected on the first and last slice of the stack (recommended).
- nuclei channel
- The number of the channel containing the nuclei.
- create results channel
- If selected a channel containing spheres, indicating the detected nuclei, is added to the stack.
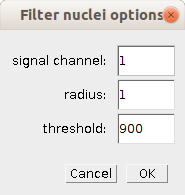
- signal channel
- The channel containing the signal by which the nuclei will be filtered.
- radius
- The radius of the circle (2D) in which the mean intensity is measured
- threshold
- The threshold value above which the nuclei are kept for the clustering
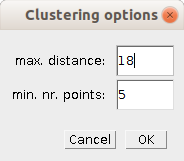
- max. distance
- The maximum distance for the DBScan-clustering algorithm [2]
- min. nr. points
- The minimum number of points a cluster must have.
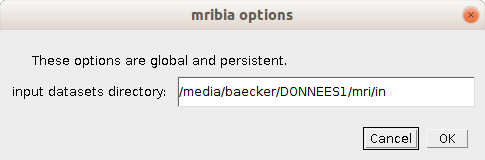
- input dataset directory
- The base directory for datasets. A sub-directory `spheroids` will be created in the base directory. The images of the dataset will be downloaded into this directory.
[1] Ollion, J., Cochennec, J., Loll, F., Escudé, C., and Boudier, T. (2013). TANGO: a generic tool for high-throughput 3D image analysis for studying nuclear organization. Bioinformatics 29, 1840–1841.
[2] Ester, M., Kriegel, H.P., Sander, J., and Xu Xiaowei. A density-based algorithm for discovering clusters in large spatial databases with noise. Proceedings of the Second International Conference on Knowledge Discovery and Data Mining (KDD-96). AAAI Press. pp. 226–231. ISBN 1-57735-004-9

 Volker Bäcker
Volker Bäcker