-
Notifications
You must be signed in to change notification settings - Fork 98
Simple example
For a simple case, imagine a bacterial genome that contains a single repeated element in two separate places in the chromosome:
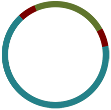
A researcher (who does not yet know the structure of the genome) sequences it, and the resulting 100 bp reads are assembled with a de novo assembler:

Because the repeated element is longer than the sequencing reads, the assembler was not able to reproduce the original genome as a single contig. Rather, three contigs are produced: one for the repeated sequence (even though it occurs twice) and one for each sequence between the repeated elements.
Given only the contigs, the relationship between these sequences is not clear. However, the assembly graph contains additional information which is made apparent in Bandage:
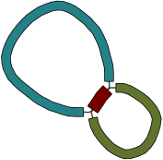
There are two principal underlying sequences compatible with this graph: two separate circular sequences that share a region in common, or a single larger circular sequence with an element that occurs twice:
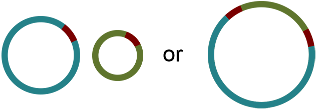
Additional knowledge, such as information on the approximate size of the bacterial chromosome, can help the researcher to rule out the first alternative. In this way, Bandage has assisted in turning a fragmented assembly of three contigs into a completed genome of one sequence.
- Home
- Getting started
- Settings:
- Functionality:
- Assembly:
- Example uses:
- Media:
- About