Instructors
Simon Prochnik
Sofia Robb
Table of Contents
- Big Picture
- Unix
- Unix 1
- Unix Overview
- The Basics
- Logging into Your Workstation
- Bringing up the Command-Line
- OK. I've Logged in. What Now?
- Command-Line Prompt
- Issuing Commands
- Command-Line Editing
- Wildcards
- Home Sweet Home
- Getting Around
- Essential Unix Commands
- Getting Information About Commands
- Finding Out What Commands are on Your Computer
- Arguments and Command Line Switches
- Spaces and Funny Characters
- Useful Commands
- Manipulating Directories
- Networking
- Standard I/O and Redirection
- A Simple Example
- Redirection Meta-Characters
- Filters, Filenames and Standard Input
- Standard I/O and Pipes
- More Pipe Idioms
- Advanced Unix
- Link to Unix 1 Problem Set
- Unix 2
- Text Editors
- Git for Beginners
- The Big Picture.
- Collaboration
- Storing Versions
- Restoring Previous Versions
- Backup
- The Details
- The Basics
- Creating a new repository
- Cloning a Repository
- Bringing Changes in Remote to your Local Repository
- Keeping track of differences between local and remote repositories
- Links to Slightly less basic topics
- Link To Unix 2 Problem Set
- Unix 1
- Python
- Python 1
- Python 2
- Python 3
- Sequences
- What functions go with my object?
- Strings
- Quotation Marks
- Strings and the print() function
- Errors and Printing
- Special/Escape Characters
- Concatenation
- The difference between string and integer
- Determine the length of a string
- Changing String Case
- Find and Count
- Find and Replace
- Extracting a Substring, or Slicing
- Locate and Report
- Other String Methods
- String Formatting
- Lists and Tuples
- Link to Python 3 Problem Set
- Python 4
- Python 5
- Python 6
- Regular Expressions
- Individual Characters
- Character Classes
- Anchors
- Quantifiers
- Variables and Patterns
- Either Or
- Subpatterns
- Using Subpatterns Inside the Regular Expression Match
- Using Subpatterns Outside the Regular Expression
- Get position of the subpattern with finditer()
- Subpatterns and Greediness
- Practical Example: Codons
- Truth and Regular Expression Matches
- Using Regular expressions in substitutions
- Using subpatterns in the replacement
- Regular Expression Option Modifiers
- Link to Python 6 Problem Set
- Regular Expressions
- Python 7
- Python 8
- Python 9
- Bioinformatic Analysis and Tools
Instructors
Simon Prochnik
Sofia Robb
Why is it important for biologists to learn to program?
You probably already know the answer to this question since you are here.
We firmly believe that knowing how to program is just as essential as knowing how to run a gel or set up a PCR reaction. The data we now get from a single experiment can be overwhelming. This data often needs to be reformatted, filtered, and analyzed in unique ways. Programming allows you to perform these tasks in an efficient and reproducible way.
What are our tips for having a successful programming course?
-
Practice, practice, practice. Please spend as much time as possible actually coding.
-
Write only a line or two of code, then test it. If you write too many lines, it becomes more difficult to debug if there is an error.
-
Errors are not failures. Every error you get is a learning opportunity. Every single error you debug is a major success. Fixing errors is how you will cement what you have learned.
-
Don't spend too much time trying to figure out a problem. While it's a great learning experience to try to solve an issue on your own, it's not fun getting frustrated or spending a lot of time stuck. We are here to help you, so please ask us whenever you need help.
-
Lectures are important, but the practice is more important.
-
Review sessions are important, but practice is more important.
-
Our key goal is to slowly, but surely, teach you how to solve problems on your own.
Underlying the pretty Mac OSX GUI is a powerful command-line operating system. The command-line gives you access to the internals of the OS, and is also a convenient way to write custom software and scripts.
Many bioinformatics tools are written to run on the command-line and have no graphical interface. In many cases, a command-line tool is more versatile than a graphical tool, because you can easily combine command-line tools into automated scripts that accomplish tasks without human intervention.
In this course, we will be writing Python scripts that are completely command-line based.
Your workstation is an iMac. To log into it, provide the following information:
Your username: admin
Your password: cshl
To bring up the command-line, use the Finder to navigate to Applications->Utilities and double-click on the Terminal application. This will bring up a window like the following:
You can open several Terminal windows at once. This is often helpful.
You will be using this application a lot, so I suggest that you drag the Terminal icon into the shortcuts bar at the bottom of your screen.
The terminal window is running shell called "bash." The shell is a loop that:
- Prints a prompt
- Reads a line of input from the keyboard
- Parses the line into one or more commands
- Executes the commands (which usually print some output to the terminal)
- Go back 1.
There are many different shells with bizarre names like bash, sh, csh, tcsh, ksh, and zsh. The "sh" part means shell. Each shell has different and somewhat confusing features. We have set up your accounts to use bash. Stay with bash and you'll get used to it, eventually.
Most of bioinformatics is done with command-line software, so you should take some time to learn to use the shell effectively.
This is a command-line prompt:
bush202>
This is another:
(~) 51%
This is another:
srobb@bush202 1:12PM>
What you get depends on how the system administrator has customized your login. You can customize it yourself when you know how.
The prompt tells you the shell is ready to accept a command. When a long-running command is going, the prompt will not reappear until the system is ready to deal with your next request.
Type in a command and press the <Enter> key. If the command has output, it will appear on the screen. Example:
(~) 53% ls -F
GNUstep/ cool_elegans.movies.txt man/
INBOX docs/ mtv/
INBOX~ etc/ nsmail/
Mail@ games/ pcod/
News/ get_this_book.txt projects/
axhome/ jcod/ public_html/
bin/ lib/ src/
build/ linux/ tmp/
ccod/
(~) 54%
The command here is ls -F which produces a listing of files and directories in the current directory (more on that later). Below its output, the command prompt appears again.
Some programs will take a long time to run. After you issue their command names, you won't recover the shell prompt until they're done. You can either launch a new shell (from Terminal's File menu), or run the command in the background by adding an ampersand after the command
(~) 54% long_running_application &
(~) 55%
The command will now run in the background until it is finished. If it has any output, the output will be printed to the terminal window. You may wish to capture the output in a file (called redirection). We'll describe this later.
Most shells offer command-line editing. Up until the comment you press , you can go back over the command-line and edit it using the keyboard. Here are the most useful keystrokes:
- Backspace: Delete the previous character and back up one.
- Left arrow, right arrow: Move the text insertion point (cursor) one character to the left or right.
- control-a (^a): Move the cursor to the beginning of the line. (Mnemonic: A is first letter of alphabet)
- control-e (^e): Move the cursor to the end of the line. Mnemonic: E for the End (^z was already used for interrupt a command).
- control-d (^d): Delete the character currently under the cursor. D=Delete.
- control-k (^k): Delete the entire line from the cursor to the end. k=kill. The line isn't actually deleted, but put into a temporary holding place called the "kill buffer". This is like cutting text
- control-y (^y): Paste the contents of the kill buffer onto the command-line starting at the cursor. y=yank. This is like paste.
- Up arrow, down arrow: Move up and down in the command history. This lets you reissue previous commands, possibly after modifying them.
There are also some useful shell commands you can issue:
historyShow all the commands that you have issued recently, nicely numbered.!<number>Reissue an old command, based on its number (which you can get fromhistory).!!Reissue the immediate previous command.!<partial command string>: Reissue the previous command that began with the indicated letters. For example,!l(the letter el, not a number 1) would reissue thels -Fcommand from the earlier example.
bash offers automatic command completion and spelling correction. If you type part of a command and then the tab key, it will prompt you with all the possible completions of the command. For example:
(~) 51% fd<tab><tab>
(~) 51% fd
fd2ps fdesign fdformat fdlist fdmount fdmountd fdrawcmd fdumount
(~) 51%
If you hit tab after typing a command, but before pressing <Enter>, bash will prompt you with a list of file names. This is because many commands operate on files.
You can use wildcards when referring to files. * stands for zero or more characters. ? stands for any single character. For example, to list all files with the extension ".txt", run ls with the wildcard pattern "*.txt"
(~) 56% ls -F *.txt
final_exam_questions.txt genomics_problem.txt
genebridge.txt mapping_run.txt
There are several more advanced types of wildcard patterns that you can read about in the tcsh manual page. For example, if you want to match files that begin with the characters "f" or "g" and end with ".txt", you can use a range of characters inside square brackets [f-g] as part of the wildcard pattern. Here's an example
(~) 57% ls -F [f-g]*.txt
final_exam_questions.txt genebridge.txt genomics_problem.txt
When you first log in, you'll be placed in a part of the system that is your personal directory, called the home directory. You are free to do with this area what you will: in particular you can create and delete files and other directories. In general, you cannot create files elsewhere in the system.
Your home directory lives somewhere in the filesystem. On our iMacs, it is a directory with the same name as your login name, located in /Users. The full directory path is therefore /Users/username. Since this is a pain to write, the shell allows you to abbreviate it as ~username (where "username" is your user name), or simply as ~. The weird character (called "tilde" or "twiddle") is usually hidden at the upper left corner of your keyboard.
To see what is in your home directory, issue the command ls -F:
(~) % ls -F
INBOX Mail/ News/ nsmail/ public_html/
This shows one file "INBOX" and four directories ("Mail", "News") and so on. (The -F in the command turns on fancy mode, which appends special characters to directory listings to tell you more about what you're seeing. / at the end of a filename means that file is a directory.)
In addition to the files and directories shown with ls -F, there may be one or more hidden files. These are files and directories whose names start with a . (called the "dot" character). To see these hidden files, add an a to the options sent to the ls command:
(~) % ls -aF
./ .cshrc .login Mail/
../ .fetchhost .netscape/ News/
.Xauthority .fvwmrc .xinitrc* nsmail/
.Xdefaults .history .xsession@ public_html/
.bash_profile .less .xsession-errors
.bashrc .lessrc INBOX
Whoa! There's a lot of hidden stuff there. But don't go deleting dot files. Many of them are essential configuration files for commands and other programs. For example, the
.profilefile contains configuration information for the bash shell. You can peek into it and see all of bash's many options. You can edit it (when you know what you're doing) in order to change things like the command prompt and command search path.
You can move around from directory to directory using the cd command. Give the name of the directory you want to move to, or give no name to move back to your home directory. Use the pwd command to see where you are (or rely on the prompt, if configured):
(~/docs/grad_course/i) 56% cd
(~) 57% cd /
(/) 58% ls -F
bin/ dosc/ gmon.out mnt/ sbin/
boot/ etc/ home@ net/ tmp/
cdrom/ fastboot lib/ proc/ usr/
dev/ floppy/ lost+found/ root/ var/
(/) 59% cd ~/docs/
(~/docs) 60% pwd
/usr/home/lstein/docs
(~/docs) 62% cd ../projects/
(~/projects) 63% ls
Ace-browser/ bass.patch
Ace-perl/ cgi/
Foo/ cgi3/
Interface/ computertalk/
Net-Interface-0.02/ crypt-cbc.patch
Net-Interface-0.02.tar.gz fixer/
Pts/ fixer.tcsh
Pts.bak/ introspect.pl*
PubMed/ introspection.pm
SNPdb/ rhmap/
Tie-DBI/ sbox/
ace/ sbox-1.00/
atir/ sbox-1.00.tgz
bass-1.30a/ zhmapper.tar.gz
bass-1.30a.tar.gz
(~/projects) 64%
Each directory contains two special hidden directories named
.and... The first,.refers always to the current directory...refers to the parent directory. This lets you move upward in the directory hierarchy like this:
(~/docs) 64% cd ..
and to do arbitrarily weird things like this:
(~/docs) 65% cd ../../lstein/docs
The latter command moves upward two levels, and then into a directory named
docsinside a directory calledlstein.
If you get lost, the pwd command prints out the full path to the current directory:
(~) 56% pwd
/Users/lstein
With the exception of a few commands that are built directly into the shell, all Unix commands are standalone executable programs. When you type the name of a command, the shell will search through all the directories listed in the PATH environment variable for an executable of the same name. If found, the shell will execute the command. Otherwise, it will give a "command not found" error.
Most commands live in /bin, /usr/bin, or /usr/local/bin.
The man command will give a brief synopsis of a command. Let's get information about the command wc
(~) 76% man wc
Formatting page, please wait...
WC(1) WC(1)
NAME
wc - print the number of bytes, words, and lines in files
SYNOPSIS
wc [-clw] [--bytes] [--chars] [--lines] [--words] [--help]
[--version] [file...]
DESCRIPTION
This manual page documents the GNU version of wc. wc
counts the number of bytes, whitespace-separated words,
...
The apropos command will search for commands matching a keyword or phrase. Here's an example that looks for commands related to 'column'
(~) 100% apropos column
showtable (1) - Show data in nicely formatted columns
colrm (1) - remove columns from a file
column (1) - columnate lists
fix132x43 (1) - fix problems with certain (132 column) graphics
modes
Many commands take arguments. Arguments are often the names of one or more files to operate on. Most commands also take command-line "switches" or "options", which fine-tune what the command does. Some commands recognize "short switches" that consist of a minus sign - followed by a single character, while others recognize "long switches" consisting of two minus signs -- followed by a whole word.
The wc (word count) program is an example of a command that recognizes both long and short options. You can pass it the -c, -w and/or -l options to count the characters, words and lines in a text file, respectively. Or you can use the longer but more readable, --chars, --words or --lines options. Both these examples count the number of characters and lines in the text file /var/log/messages:
(~) 102% wc -c -l /var/log/messages
23 941 /var/log/messages
(~) 103% wc --chars --lines /var/log/messages
23 941 /var/log/messages
You can cluster short switches by concatenating them together, as shown in this example:
(~) 104% wc -cl /var/log/messages
23 941 /var/log/messages
Many commands will give a brief usage summary when you call them with the -h or --help switch.
The shell uses whitespace (spaces, tabs and other non-printing characters) to separate arguments. If you want to embed whitespace in an argument, put single quotes around it. For example:
mail -s 'An important message' 'Bob Ghost <[email protected]>'
This will send an e-mail to the fictitious person Bob Ghost. The -s switch takes an argument, which is the subject line for the e-mail. Because the desired subject contains spaces, it has to have quotes around it. Likewise, my name and e-mail address, which contains embedded spaces, must also be quoted in this way.
Certain special non-printing characters have escape codes associated with them:
| Escape Code | Description |
|---|---|
| \n | new line character |
| \t | tab character |
| \r | carriage return character |
| \a | bell character (ding! ding!) |
| \nnn | the character whose ASCII code is nnn |
Here are some commands that are used extremely frequently. Use man to learn more about them. Some of these commands may be useful for solving the problem set ;-)
| Command | Description |
|---|---|
ls |
Directory listing. Most frequently used as ls -F (decorated listing), ls -l (long listing), ls -a (list all files). |
mv |
Rename or move a file or directory. |
cp |
Copy a file. |
rm |
Remove (delete) a file. |
mkdir |
Make a directory |
rmdir |
Remove a directory |
ln |
Create a symbolic or hard link. |
chmod |
Change the permissions of a file or directory. |
| Command | Description |
|---|---|
cat |
Concatenate program. Can be used to concatenate multiple files together into a single file, or, much more frequently, to view the contents of a file or files in the terminal. |
echo |
print a copy of some text to the screen. E.g. echo 'Hello World!' |
more |
Scroll through a file page by page. Very useful when viewing large files. Works even with files that are too big to be opened by a text editor. |
less |
A version of more with more features. |
head |
View the first few lines of a file. You can control how many lines to view. |
tail |
View the end of a file. You can control how many lines to view. You can also use tail -f to view a file that you are writing to. |
wc |
Count words, lines and/or characters in one or more files. |
tr |
Substitute one character for another. Also useful for deleting characters. |
sort |
Sort the lines in a file alphabetically or numerically. |
uniq |
Remove duplicated lines in a file. |
cut |
Remove columns from each line of a file or files. |
fold |
Wrap each input line to fit in a specified width. |
grep |
Filter a file for lines matching a specified pattern. Can also be reversed to print out lines that don't match the specified pattern. |
gzip (gunzip) |
Compress (uncompress) a file. |
tar |
Archive or unarchive an entire directory into a single file. |
emacs |
Run the Emacs text editor (good for experts). |
vi |
Run the vi text editor (better for experts). |
| Command | Description |
|---|---|
ssh |
A secure (encrypted) way to log into machines. |
scp |
A secure way to copy (cp) files to and from remote machines. |
ping |
See if a remote host is up. |
ftp/ sftp (secure) |
transfer files using the File Transfer Protocol. |
Unix commands communicate via the command-line interface. They can print information out to the terminal for you to see, and accept input from the keyboard (that is, from you!)
Every Unix program starts out with three connections to the outside world. These connections are called "streams", because they act like a stream of information (metaphorically speaking):
| Stream Type | Description |
|---|---|
| standard input | This is a communications stream initially attached to the keyboard. When the program reads from standard input, it reads whatever text you type in. |
| standard output | This stream is initially attached to the terminal. Anything the program prints to this channel appears in your terminal window. |
| standard error | This stream is also initially attached to the terminal. It is a separate channel intended for printing error messages. |
The word "initially" might lead you to think that standard input, output and error can somehow be detached from their starting places and reattached somewhere else. And you'd be right. You can attach one or more of these three streams to a file, a device, or even to another program. This sounds esoteric, but it is actually very useful.
The wc program counts lines, characters and words in data sent to its standard input. You can use it interactively like this:
(~) 62% wc
Mary had a little lamb,
little lamb,
little lamb.
Mary had a little lamb,
whose fleece was white as snow.
^D
6 20 107
In this example, I ran the wc program. It waited for me to type in a little poem. When I was done, I typed the END-OF-FILE character, control-d (^d for short). wc then printed out three numbers indicating the number of lines, words and characters in the input.
More often, you'll want to count the number of lines in a big file; say a file filled with DNA sequences. You can do this by redirecting the contents of a file to the standard input of wc. This uses
the < symbol:
(~) 63% wc < big_file.fasta
2943 2998 419272
If you wanted to record these counts for posterity, you could redirect standard output as well using the > symbol:
(~) 64% wc < big_file.fasta > count.txt
Now if you cat the file count.txt, you'll see that the data has been recorded. cat works by taking its standard input and copying it to standard output. We redirect standard input from the count.txt file, and leave standard output at its default, attached to the terminal:
(~) 65% cat < count.txt
2943 2998 419272
Here's the complete list of redirection commands for bash:
| Redirect command | Description |
|---|---|
< myfile.txt |
Redirect the contents of the file to standard input |
> myfile.txt |
Redirect standard output to file |
>> logfile.txt |
Append standard output to the end of the file |
1 > myfile.txt |
Redirect just standard output to file (same as above) |
2 > myfile.txt |
Redirect just standard error to file |
> myfile.txt 2>&1 |
Redirect both stdout and stderr to file |
These can be combined. For example, this command redirects standard input from the file named /etc/passwd, writes its results into the file search.out, and writes its error messages (if any) into a file named search.err. What does it do? It searches the password file for a user named "root" and returns all lines that refer to that user.
(~) 66% grep root < /etc/passwd > search.out 2> search.err
Many Unix commands act as filters, taking data from a file or standard input, transforming the data, and writing the results to standard output. Most filters are designed so that if they are called with one or more filenames on the command-line, they will use those files as input. Otherwise they will act on standard input. For example, these two commands are equivalent:
(~) 66% grep 'gatttgc' < big_file.fasta
(~) 67% grep 'gatttgc' big_file.fasta
Both commands use the grep command to search for the string "gatttgc" in the file big_file.fasta. The first one searches standard input, which happens to be redirected from the file. The second command is explicitly given the name of the file on the command line.
Sometimes you want a filter to act on a series of files, one of which happens to be standard input. Many commands let you use - on the command-line as an alias for standard input. Example:
(~) 68% grep 'gatttgc' big_file.fasta bigger_file.fasta -
This example searches for "gatttgc" in three places. First it looks in file big_file.fasta, then in bigger_file.fasta, and lastly in standard input (which, since it isn't redirected, will come from the keyboard).
The coolest thing about the Unix shell is its ability to chain commands together into pipelines. Here's an example:
(~) 65% grep gatttgc big_file.fasta | wc -l
22
There are two commands here. grep searches a file or standard input for lines containing a particular string. Lines which contain the string are printed to standard output. wc -l is the familiar word count program, which counts words, lines and characters in a file or standard input. The -l command-line option instructs wc to print out just the line count. The | character, which is known as a "pipe", connects the two commands together so that the standard output of grep becomes the standard input of wc. Think of pipes connecting streams of data flowing.
What does this pipe do? It prints out the number of lines in which the string "gatttgc" appears in the file big_file.fasta.
Pipes are very powerful. Here are some common command-line idioms.
Count the Number of Times a Pattern does NOT Appear in a File
The example at the top of this section showed you how to count the number of lines in which a particular string pattern appears in a file. What if you want to count the number of lines in which a pattern does not appear?
Simple. Reverse the test with the -v switch:
(~) 65% grep -v gatttgc big_file.fasta | wc -l
2921
Uniquify Lines in a File
If you have a long list of names in a text file, and you want to weed out the duplicates:
(~) 66% sort long_file.txt | uniq > unique.out
This works by sorting all the lines alphabetically and piping the result to the uniq program, which removes duplicate lines that occur one after another. That's why you need to sort first. The output is placed in a file named unique.out.
Concatenate Several Lists and Remove Duplicates
If you have several lists that might contain repeated entries among them, you can combine them into a single unique list by concatenating them together, then sorting and uniquifying them as before:
(~) 67% cat file1 file2 file3 file4 | sort | uniq
Count Unique Lines in a File
If you just want to know how many unique lines there are in the file, add a wc to the end of the pipe:
(~) 68% sort long_file.txt | uniq | wc -l
Page Through a Really Long Directory Listing
Pipe the output of ls to the more program, which shows a page at a time. If you have it, the less program is even better:
(~) 69% ls -l | more
Monitor a Growing File for a Pattern
Pipe the output of tail -f (which monitors a growing file and prints out the new lines) to grep. For example, this will monitor the /var/log/syslogfile for the appearance of e-mails addressed to 'mzhang':
(~) 70% tail -f /var/log/syslog | grep mzhang
Here are a few more advanced Unix commands that are very useful and when you have time you should investigate further. We list the page numbers in the Internet Version (v3) of 'The Linux Command Line' by William Shotts.
awksed(p.295)perlone-linersforloops (p. 453)
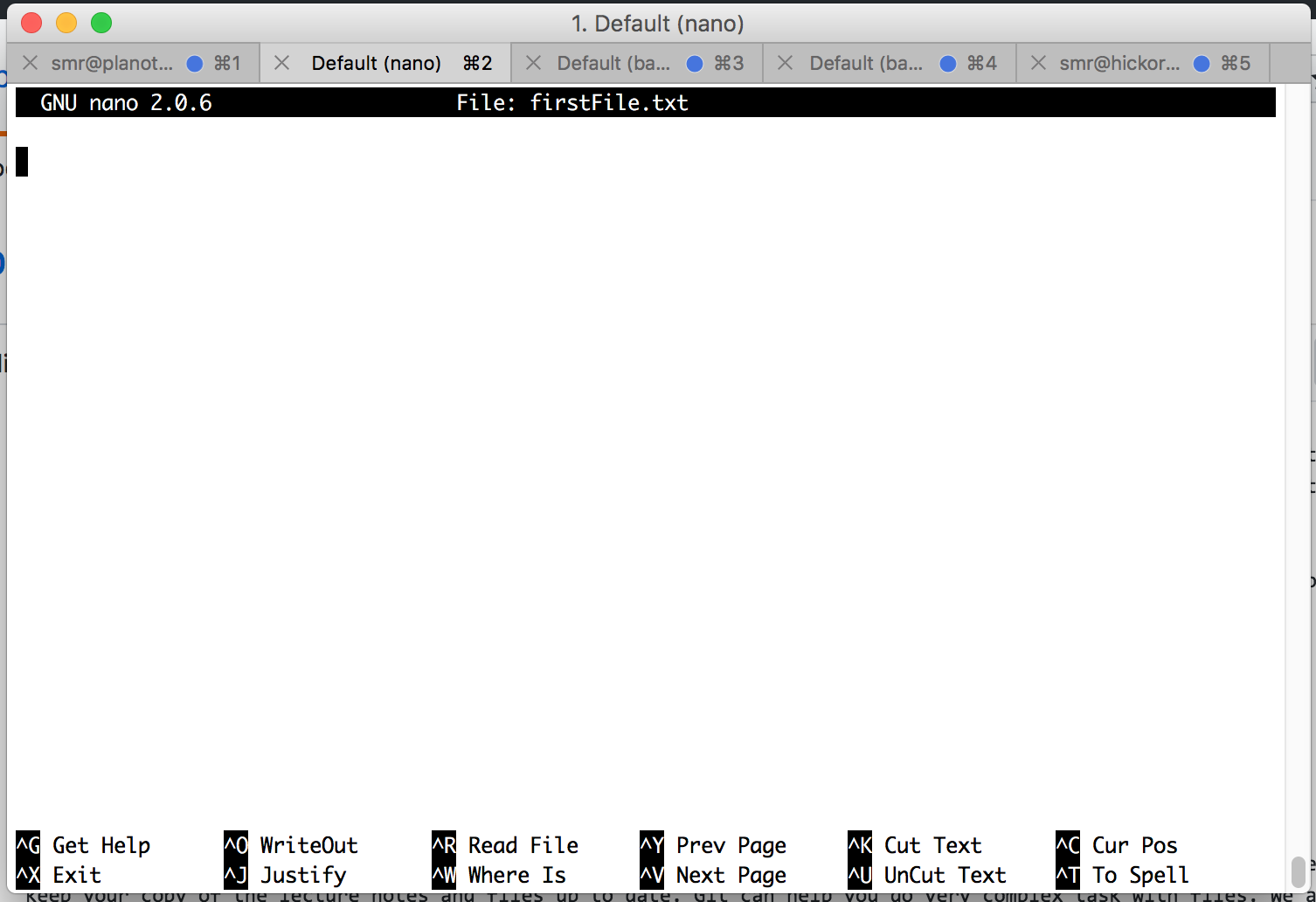
It is often necessary to create and write to a file while using the terminal. This makes it essential to use a terminal text editor. There are many text editors out there. Some of our favorite are Emacs and vim. We are going to start you out with a simple text editor called nano
The way you use nano to create a file is simply by typing the command nano followed by the name of the file you wish to create.
(~) 71% nano firstFile.txt
This is what you will see:
Things to notice:
- At the top
- the name of the program (nano) and its version number
- the name of the file you’re editing
- and whether the file has been modified since it was last saved.
- In the middle
- you will see either a blank area or text you have typed
- At the bottom
- A listing of keyboard commands such as Save (control + o) and Exit (control + x)
Keyboard commands are the only way to interact with the editor. You cannot use your mouse or trackpad.
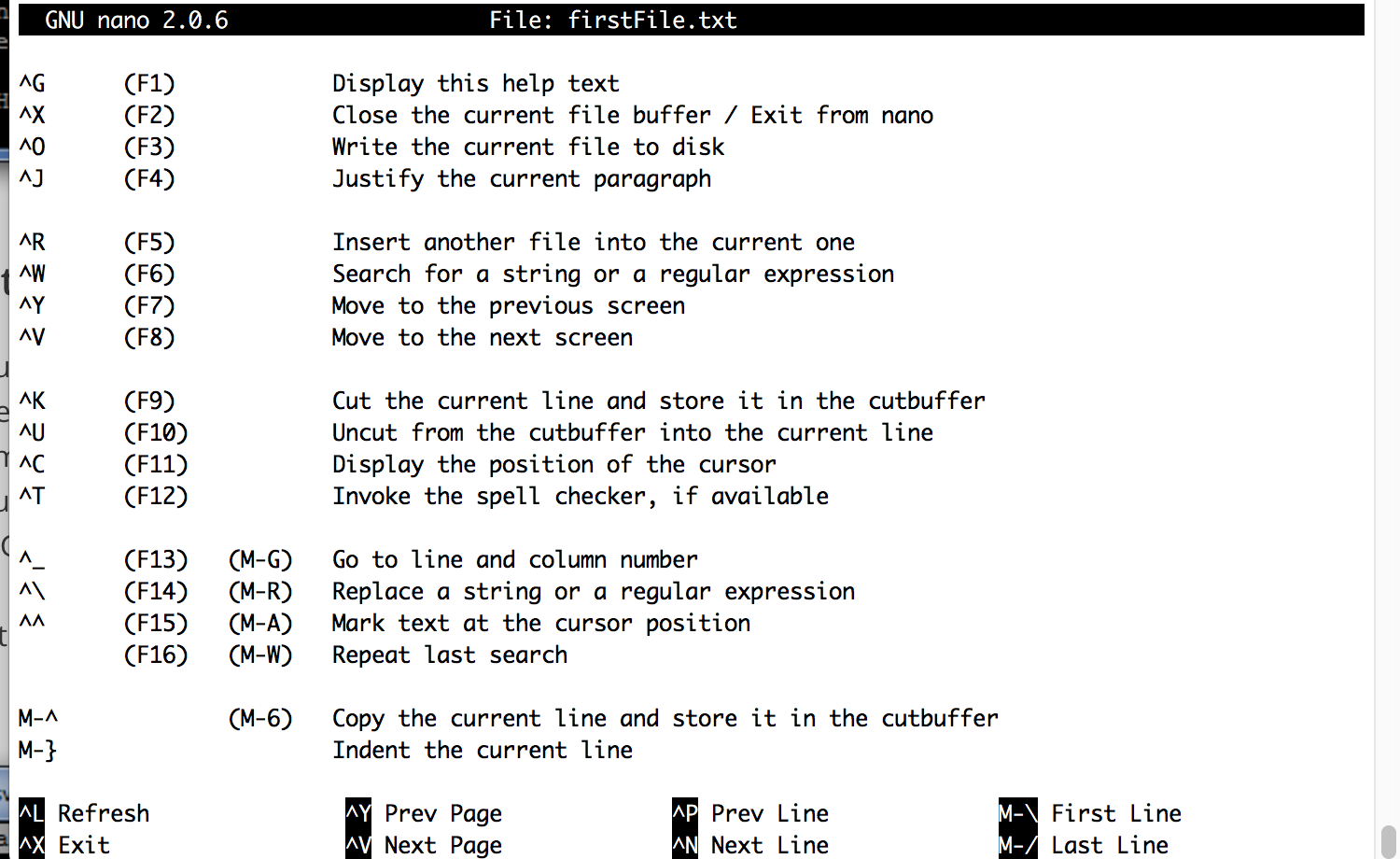
Find more commands by using control g:
The Meta key is <esc>. To use the Meta+key, hit <esc>, release, then hit the following key
Helpful commands:
- Jump to a specific line:
- control + _ then line number
- Copy a block of highlighted text
- control + ^ then move your cursor to start to highlight a block for copying
- Meta + ^ to end your highlight block
- Paste
- control + u
Nano is a beginner's text editor. vi and Emacs are better choices once you become a bit more comfortable using the terminal. These editors do cool stuff like syntax highlighting.
Git is a tool for managing files and versions of files. It is a Version Control System. It allows you to keep track of changes. You are going to be using Git to manage your course work and keep your copy of the lecture notes and files up to date. Git can help you do very complex task with files. We are going to keep it simple.
A Version Control System is good for Collaborations, Storing Versions, Restoring Previous Versions, and Managing Backups.
Using a Version Control System makes it possible to edit a document with others without the fear of overwriting someone's changes, even if more than one person is working on the same part of the document. All the changes can be merged into one document. These documents are all stored one place.
A Version Control System allows you to save versions of your files and to attach notes to each version. Each save will contain information about the lines that were added or alted.
Since you are keeping track of versions, it is possible to revert all the files in a project or just one file to a previous version.
A Version Control System makes it so that you work locally and sync your work remotely. This means you will have a copy of your project on your computer and the Version Control System Server you are using.
git is the Version Control System we will be using for tracking changes in our files.
GitHub is the Version Control System Server we will be using. They provide free account for all public projects.
A repository is a project that contains all of the project files, and stores each file's revision history. Repositories can have multiple collaborators. Repositories usually have two components, one remote and one local.
Let's Do It!
Follow Steps 1 and 2 to create the remote repository. Follow Step 3 to create your local repository and link it to the remote.
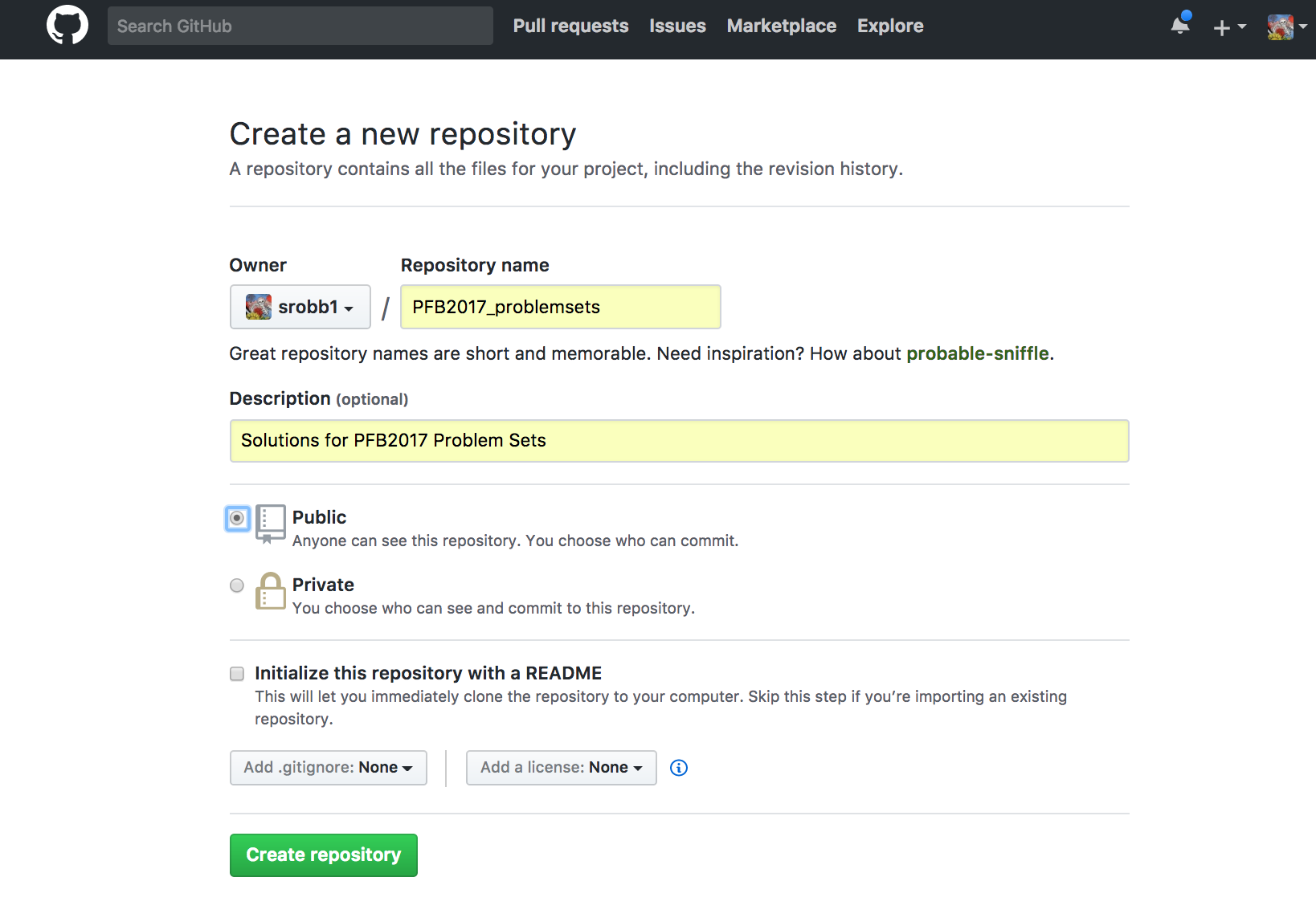
- Navigate to GitHub --> Create Account / Log In --> Go To Repositories --> Click 'New'
- Add a name (i.e., PFB2017_problemsets) and a description (i.e., Solutions for PFB2017 Problem Sets) and click "Create Repository"
- Create a directory on your computer and follow the instructions provided.
- Open your terminal and navigate to the location you want to put a directory for your problem sets
- Create a new directory directory (i.e., PFB2017_problemsets)
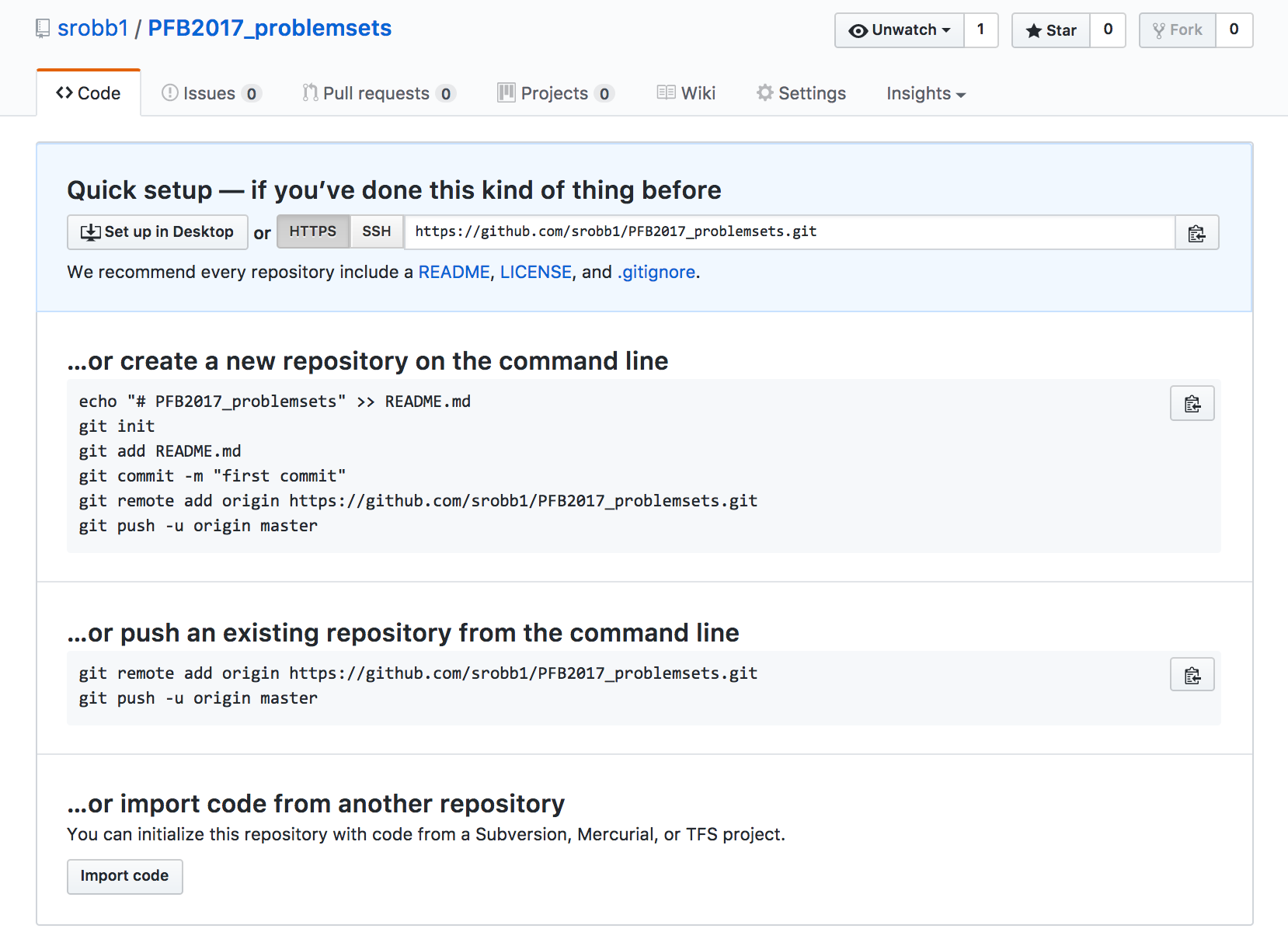
- Follow the instructions provided when you created your repository. These are my instructions, yours will be different.
echo "# PFB2017_problemsets" >> README.md
git init
git add README.md
git commit -m "first commit"
git remote add origin https://github.com/prog4biol/PFB2017_problemsets.git
git push -u origin master
You now have a repository!
Let's back up a bit and talk more about git and about these commands. For basic git use, these are almost all the command you will need to know.
Every git repository has three main elements called trees:
- The Working Directory contains your files
- The Index is the staging area
- The HEAD points to the last commit you made.
There are a few new words here, we will explain them as we go
| command | description |
|---|---|
git init |
Creates your new local repository with the three trees on (local machine) |
git remote add remote-name URL |
Links your local repository to a remote repository that is often named origin and is found at the given URL. |
git add filename |
Propose changes and add file(s) with changes to the index or staging area (local machine) |
git commit -m 'message' |
Confirm or commit that you really want to add your changes to the HEAD (local machine) |
git push -u remote-name remote-branch |
Upload your committed changes in the HEAD to the specified remote repository to the specified branch |
Let's Do it!
- Make sure you are in your local repository
- Create a new file with nano:
nano git_exercises.txt - Add a line of text to the new file.
- Save (control + o) and Exit (control + x)
- (Add) Stage your changes.
git add git_exercises.txt - (Commit) Become sure you want your changes your changes.
git commit -m 'added a line of text' - (Push) Sync/Upload your changes to the remote repository.
git push origin master
That is all there is to it! There are more complicated things you can do but we won't get into those. You will know when you are ready to learn more about git when you figure out there is something you want to do but don't know how. There are thousands of online tutorials for you to search and follow.
Sometimes you want to download and use someone else's repository.
Let's clone the course material.
Let's do it!
- Go to our PFB2017 GitHub Repository
- Click the 'Clone or Download' Button
- Copy the URL ~Clone PFB2017
- Clone the repository to your local machine
git clone https://github.com/prog4biol/pfb2017.git
Now you have a copy of the course material on your computer!
If we make changes to any of these files and you want to update your copy you can pull the changes.
git pull
If you are ever wondering what do you need to add to your remote repository use the git status command. This will provide you a list of file that have been modified, deleted, and those that are untracked. Untracked files are those that have never been added to the staging area with git add
You will KNOW if you need to use these features of git.
Python is a scripting language. It is useful for writing medium-sized scientific coding projects. When you run a Python script, the Python program will generate byte code and interpret the byte code. This happens automatically and you don't have to worry about it. Compiled languages like C, C++ will run much faster, but are much much more complicated to program. Languages like java (which also gets compiled into byte code) are well suited to very large collaborative programming projects, but don't run as fast as C and are more complex that Python.
Python has
- data types
- functions
- objects
- classes
- methods
Data types are just different types of data which are discussed in more detail later. Examples of data types are integer numbers and strings of letters and numbers (text). These can be stored in variables.
Functions do something with data, such as a calculation. Some functions are already built into Python. You can create your own functions as well.
Objects are a way of grouping a set of data and functions (methods) that act on that data
Classes are a way to encapsulate (organize) variables and functions. Objects get their variables and methods from the class they belong to.
Methods are just functions that belong to a class. Objects that belong to the a class can use methods from that class.
There are two versions of Python: Python 2 and Python 3. We will be using 3. This version fixes some of the problems with Python 2 and breaks some other things. A lot of code has already been written for Python 2 (it's older), but going forwards, more and more new code development will use Python 3.
Python can be run one line at a time in an interactive interpreter. You can think of this as a Python shell. To launch the interpreter type the following into your terminal window:
$ python3
Note: '$' indicates the command line prompt. Recall from Unix 1 that every computer can have a different prompt!
First Python Commands:
>>> print("Hello, PFB2017!")
Hello, PFB2017!Note:
print()
- The same code from above is typed into a text file using a text editor.
- Python scripts are always saved in files whose names have the extension '.py' (i.e. the filename ends with '.py').
File Contents:
print ("Hello, PFB2017!")Typing the Python command followed by the name of a script makes Python execute the script. Recall that we just saw you can run an interactive interpreter by just typing python on the command line
Execute the Python script like this (% represents the prompt)
% python3 test.py This produces the following result:
Hello, PFB2017!If you make your script executable, you can run it without typing python3 first. Use chmod to change the permissions on the script like this
chmod +x test.py
You can look at the permissions with
% ls -l test.py
-rwxr-xr-x 1 sprochnik staff 60 Oct 16 14:29 test.py
The first field of -, r, w and x characters define the permissions of the file. The three 'x' characters means anyone can execute or run this script.
We also need to add a line at the beginning of the script that tells the shell to run python3 to interpret the script. This line starts with #, so it looks like a comment to python. The '!' is important as is the space between env and python3. The program /usr/bin/env looks for where python3 is installed and runs the script with python3. The details may seem a bit complex, but you can just copy and paste this 'magic' line.
The file test.py now looks like this
#!/usr/bin/env python3
print ("Hello, PFB2017!")Now you can simply type the name of the script to run it. Like this
% ./test.py
Hello, PFB2017!
A Python identifier is a name used to identify a variable, function, class, module or other object. An identifier starts with a letter A to Z or a to z or an underscore (_) followed by zero or more letters, underscores and digits (0 to 9).
Python does not allow punctuation characters such as @, $, and % within identifiers. Python is a case sensitive programming language. Thus, seq_id and seq_ID are two different identifiers in Python.
- The first character is lowercase, unless it is a name of a class. Classes should begin with an uppercase characters. (ex. Seq)
- Private identifiers begin with an underscore. (ex.
_private) - Strong private identifiers begin with two underscores. (ex.
__private) - Language-defined special names begin and end with two underscores. (ex.
__special__)
The following is a list of Python keywords. These are special words that already have a purpose in python and therefore cannot be used as identifier names.
and exec not
as finally or
assert for pass
break from print
class global raise
continue if return
def import try
del in while
elif is with
else lambda yield
except
Python denotes blocks of code by line indentation. Incorrect line spacing and/or indention will cause an error or could make your code run in a way you don't expect. You can get help with indentation from good text editors or Interactive Development Environments (IDEs).
The number of spaces in the indentation need to be consistent but a specific number is not required. All lines of code, or statements, within a single block must be indented in the same way. For example:
#!/usr/bin/env python3
for x in (1,2,3,4,5):
if x > 4:
print("Hello")
else:
print(x)
print('All Done!')Comments are an essential programming practice. Making a note of what a line or block of code is doing will help the writer and readers of the code. This includes you!
Comments start with a pound or hash symbol #. All characters after this symbol, up to the end of the line are part of the comment and are ignored by Python.
The first line of a script starting with #! is a special example of a comment that also has the special function in unix of telling the unix shell how to run the script.
#!/usr/bin/env python3
# this is my first script
print ("Hello, PFB2017!") # this line prints output to the screenBlank lines are also important for increasing the readability of the code. You should separate pieces of code that go together with a blank line to make 'paragraphs' of code. Blank lines are ignored by the Python interpretor.
This is our first look at variables and data types. Each data type will be discussed in more detail in subsequent sections.
The first concept to consider is that Python data types are either immutable (unchangeable) or not. Literal numbers, strings and tuples cannot be changed. Lists, dictionaries and sets can be changed. So can individual (scalar) variables. You can store data in memory by putting it in different kinds variables. You use the = sign to assign a value to a variable.
Numbers and strings are two common data types. Literal numbers and strings like this 5 or 'my name is' are immutable. However, their values can be stored in variables and then changed.
For Example:
gene_count = 5
gene_count = 10You should give your variables names that help you understand what they store. gene_count, expression, sequences are all good identifiers or variable names. k, x, data, var1, var2 are bad because you can't tell what they store. This means it's harder to understand the script and to spot errors or bugs in your script.
Different types of data can be assigned to variables, i.e., integers (1,2,3), floats (floating point numbers, 3.1415), and strings (text).
For Example:
count = 10 # this is an integer
average = 2.5 # this is a float
message = "Welcome to Python" # this is a string10, 2.5, and "Welcome to Python" are singular pieces of data being stored in an individual variables.
Collections of data can also be stored in special data types, i.e., tuples, lists, sets, and dictionaries. Generally, you should try to store like with like, so each element in the data type should be the same kind of data, like an expression value from RNA-seq or a count of how many exons are in a gene or a read sequence.
- Tuples are similar to lists and contain ordered, indexed collection of data.
- Tuples are immutable: you can't change the values or the number of values
- A tuple is enclosed in parentheses and items are separated by commas.
( 'Jan' , 'Feb' , 'Mar' , 'Apr' , 'May' , 'Jun' , 'Jul' , 'Aug' , 'Sep' , 'Oct' , 'Nov' , 'Dec' )| Index | Value |
|---|---|
| 0 | Jan |
| 1 | Feb |
| 2 | Mar |
| 3 | Apr |
| 4 | May |
| 5 | Jun |
| 6 | Jul |
| 7 | Aug |
| 8 | Sep |
| 9 | Oct |
| 10 | Nov |
| 11 | Dec |
-
Dictionaries are good for storing data that can be represented as a two-column table.
-
They store unordered collections of key/value pairs.
-
A dictionary is enclosed in curly braces. and sets of Key/Value pairs are separated by commas
-
A colon is written between each key and value. Commas separate key:value pairs.
{ 'TP53' : 'GATGGGATTGGGGTTTTCCCCTCCCATGTGCTCAAGACTGGCGCTAAAAGTTTTGAGCTTCTCAAAAGTC' , 'BRCA1' : 'GTACCTTGATTTCGTATTCTGAGAGGCTGCTGCTTAGCGGTAGCCCCTTGGTTTCCGTGGCAACGGAAAA' }| Key | Value |
|---|---|
| TP53 | GATGGGATTGGGGTTTTCCCCTCCCATGTGCTCAAGACTGGCGCTAAAAGTTTTGAGCTTCTCAAAAGTC |
| BRCA1 | GTACCTTGATTTCGTATTCTGAGAGGCTGCTGCTTAGCGGTAGCCCCTTGGTTTCCGTGGCAACGGAAAA |
- Lists are used to store an ordered, indexed collection of data.
- Lists are mutable: the number of elements in the list and what's stored in each element can change
- Lists are enclosed in square brackets and items are separated by commas
[ 'atg' , 'aaa' , 'agg' ]| Index | Value |
|---|---|
| 0 | atg |
| 1 | aaa |
| 2 | agg |
The list index starts at 0
Command line parameters follow the name of a script or program and have spaces between them. They allow a user to pass information to a script on the command line when that script is being run. Python stores all the pieces of the command line in a special list called sys.argv.
You need to import the sys module at the beginning of your script like this
#!/usr/bin/env python3
import sysLet's imagine we have a script called friends.py. If you write this on the command line:
$ friends.py Joe AnitaThis happens inside the script:
the script name 'friends.py', and the strings 'Joe' and 'Anita' appear in a list called
sys.argv.
These are the command line parameters, or arguments you want to pass to your script.
sys.argv[0]is the script name.
You can access values of the other parameters by their indices, starting with 1, sosys.argv[1]contains 'Joe' andsys.argv[2]contains 'Anita'. You access elements in a list by adding square brackets and the numerical index after the name of the list. If you wanted to print a message saying these two people are friends, you might write some code like this
#!/usr/bin/env python3
import sys
friend1 = sys.argv[1] # get first command line parameter
friend2 = sys.argv[2] # get second command line parameter
# now print a message to the screen
print(friend1,'and',friend2,'are friends')The advantage of getting input from the user from the command line is that you can write a script that is general. It can print a message with any input the user provides. This makes it flexible. The user also supplies all the data the script needs on the command line so the script doesn't have to ask the user to input a name and wait til the user does this. The script can run on its own with no further interaction from the user. This frees the user to work on something else. Very handy!
You have an identifier in your code called data. Does it represent a string or a list or a dictionary? Python has a couple of functions that help you figure this out.
| Function | Description |
|---|---|
type(data) |
tells you which class your object belongs to |
dir(data) |
tells you which methods are available for your object |
id(data) |
tells you the unique object id |
We'll cover dir() in more detail later
>>> data = [2,4,6]
>>> type(data)
<class 'list'>
>>> data = 5
>>> type(data)
<class 'int'>An operator in a programming language is a symbol that tells the compiler or interpreter to perform specific mathematical, relational or logical operation and produce a result. Here we explain the concept of operators.
In Python we can write statements that perform mathematical calculations. To do this we need to use operators that are specific for this purpose. Here are arithmetic operators:
| Operator | Description | Example | Result |
|---|---|---|---|
+ |
Addition | 3+2 |
5 |
- |
Subtraction | 3-2 |
1 |
* |
Multiplication | 3*2 |
6 |
/ |
Division | 3/2 |
1.5 |
% |
Modulus (divides left operand by right operand and returns the remainder) | 3%2 |
1 |
** |
Exponent | 3**2 |
9 |
// |
Floor Division (result is the quotient with digits after the decimal point removed. If one of the operands is negative, the result is floored, i.e., rounded away from zero | 3//2 ; -11//3 |
1 ; -4 |
We use assignment operators to assign values to variables. You have been using the = assignment operator. Here are others:
| Operator | Equivalent to | Example | result evaluates to |
|---|---|---|---|
= |
a = 3 |
result = 3 |
3 |
+= |
result = result + 2 |
result = 3 ; result += 2 |
5 |
-= |
result = result - 2 |
result = 3 ; result -= 2 |
1 |
*= |
result = result * 2 |
result = 3 ; result *= 2 |
6 |
/= |
result = result / 2 |
result = 3 ; result /= 2 |
1.5 |
%= |
result = result % 2 |
result = 3 ; result %= 2 |
1 |
**= |
result = result ** 2 |
result = 3 ; result **= 2 |
9 |
//= |
result = result // 2 |
result = 3 ; result //= 3 |
1 |
These operators compare two values and returns true or false.
| Operator | Description | Example | Result |
|---|---|---|---|
== |
equal to | 3 == 2 |
False |
!= |
not equal | 3 != 2 |
True |
> |
greater than | 3 > 2 |
True |
< |
less than | 3 < 2 |
False |
>= |
greater than or equal | 3 >= 2 |
True |
<= |
less than or equal | 3 <= 2 |
False |
Logical operators allow you to combine two or more sets of comparisons. You can combine the results in different ways. For example you can 1) demand that all the statements are true, 2) that only one statement needs to be true, or 3) that the statement needs to be false.
| Operator | Description | Example | Result |
|---|---|---|---|
and |
True if left operand is True and right operand is True | 3>=2 and 2<3 |
True |
or |
True if left operand is True or right operand is True | 3==2 or 2<3 |
True |
not |
Reverses the logical status | not False |
True |
You can test to see if a value is included in a string, tuple, or list. You can also test that the value is not included in the string, tuple, or list.
| Operator | Description |
|---|---|
in |
True if a value is included in a list, tuple, or string |
not in |
True if a value is absent in a list, tuple, or string |
For Example:
>>> dna = 'GTACCTTGATTTCGTATTCTGAGAGGCTGCTGCTTAGCGGTAGCCCCTTGGTTTCCGTGGCAACGGAAAA'
>>> 'TCT' in dna
True
>>>
>>> 'ATG' in dna
False
>>> 'ATG' not in dna
True
>>> codons = [ 'atg' , 'aaa' , 'agg' ]
>>> 'atg' in codons
True
>>> 'ttt' in codons
FalseOperators are listed in order of precedence. Highest listed first. Not all the operators listed here are mentioned above.
| Operator | Description |
|---|---|
** |
Exponentiation (raise to the power) |
~ + - |
Complement, unary plus and minus (method names for the last two are +@ and -@) |
* / % // |
Multiply, divide, modulo and floor division |
+ - |
Addition and subtraction |
>> << |
Right and left bitwise shift |
& |
Bitwise 'AND' |
^ | |
Bitwise exclusive 'OR' and regular 'OR' |
<= < > >= |
Comparison operators |
<> == != |
Equality operators |
= %= /= //= -= += *= **= |
Assignment operators |
is |
Identity operator |
is not |
Non-identity operator |
in |
Membership operator |
not in |
Negative membership operator |
not or and |
logical operators |
Note: Find out more about bitwise operators. We will see these operators used in the section on Sets.
Lets take a step back, What is truth?
Everything is true, except for:
| expression | TRUE/FALSE |
|---|---|
0 |
FALSE |
None |
FALSE |
False |
FALSE |
'' (empty string) |
FALSE |
[] (empty list) |
FALSE |
() (empty tuple) |
FALSE |
{} (empty dictionary) |
FALSE |
Which means that these are True:
| expression | TRUE/FALSE |
|---|---|
'0' |
TRUE |
'None' |
TRUE |
'False' |
TRUE |
'True' |
TRUE |
' ' (string of one blank space) |
TRUE |
bool() is a function that will test if a value is true.
>>> bool(True)
True
>>> bool('True')
True
>>>
>>>
>>> bool(False)
False
>>> bool('False')
True
>>>
>>>
>>> bool(0)
False
>>> bool('0')
True
>>>
>>>
>>> bool('')
False
>>> bool(' ')
True
>>>
>>>
>>> bool(())
False
>>> bool([])
False
>>> bool({})
FalseControl Statements are used to direct the flow of your code and create the opportunity for decision making. Control statements foundation is build on truth.
- Use the
ifStatement to test for truth and to execute lines of code if true. - When the expression evaluates to true each of the statements indented below the
ifstatment, also known as the nested statement block, will be executed.
if
if expression :
statement
statementFor Example:
dna = 'GTACCTTGATTTCGTATTCTGAGAGGCTGCTGCTTAGCGGTAGCCCCTTGGTTTCCGTGGCAACGGAAAA'
if 'AGC' in dna:
print('found AGC in your dna sequence')Returns:
found AGC in your dna sequence
else
- The
ifportion of the if/else statement behave as before. - The first indented block is executed if the condition is true.
- If the condition is false, the second indented else block is executed.
dna = 'GTACCTTGATTTCGTATTCTGAGAGGCTGCTGCTTAGCGGTAGCCCCTTGGTTTCCGTGGCAACGGAAAA'
if 'ATG' in dna:
print('found ATG in your dna sequence')
else:
print('did not find ATG in your dna sequence')Returns:
did not find ATG in your dna sequence
- The if condition is tested as before and the indented block is executed if the condition is true.
- If it's false, the indented block following the elif is executed if the first elif condition is true.
- Any remaining elif conditions will be tested in order until one is found to be true. If none is true, the else indented block is executed.
count = 60
if count < 0:
message = "is less than 0"
print(count, message)
elif count < 50:
message = "is less than 50"
print (count, message)
elif count > 50:
message = "is greater than 50"
print (count, message)
else:
message = "must be 50"
print(count, message)Returns:
60 is greater than 50
Let's change count to 20, which statement block gets executed?
count = 20
if count < 0:
message = "is less than 0"
print(count, message)
elif count < 50:
message = "is less than 50"
print (count, message)
elif count > 50:
message = "is greater than 50"
print (count, message)
else:
message = "must be 50"
print(count, message)Returns:
20 is less than 100
What happens when count is 50?
count = 50
if count < 0:
message = "is less than 0"
print(count, message)
elif count < 50:
message = "is less than 50"
print (count, message)
elif count > 50:
message = "is greater than 50"
print (count, message)
else:
message = "must be 50"
print(count, message)Returns:
50 must be 50
Python recognizes 3 types of numbers: integers, floating point numbers, and complex numbers.
- known as an int
- an int can be positive or negative
- and does not contain a decimal point or exponent.
- known as a float
- a floating point number can be positive or negative
- and does contain a decimal point (
4.875) or exponent (4.2e-12)
- known as complex
- is in the form of a+bi where bi is the imaginary part.
Sometimes one type of number needs to be changed to another for a function to be able to do work on it. Here are a list of functions for converting number types:
| function | Description |
|---|---|
int(x) |
to convert x to a plain integer |
float(x) |
to convert x to a floating-point number |
complex(x) |
to convert x to a complex number with real part x and imaginary part zero |
complex(x, y) |
to convert x and y to a complex number with real part x and imaginary part y |
>>> int(2.3)
2
>>> float(2)
2.0
>>> complex(2.3)
(2.3+0j)
>>> complex(2.3,2)
(2.3+2j)Here are a list of functions that take numbers as arguments. These use useful things like rounding.
| function | Description |
|---|---|
abs(x) |
The absolute value of x: the (positive) distance between x and zero. |
round(x [,n]) |
x rounded to n digits from the decimal point. round() rounds to an even integer if the value is exactly between two integers, so round(0.5) is 0 and round(-0.5) is 0. round(1.5) is 2. Rounding to a fixed number of decimal places can give unpredictable results. |
max(x1, x2,...) |
The largest positive argument is returned |
min(x1, x2,...) |
The smallest argument is returned |
>>> abs(2.3)
2.3
>>> abs(-2.9)
2.9
>>> round(2.3)
2
>>> round(2.5)
2
>>> round(2.9)
3
>>> round(-2.9)
-3
>>> round(-2.3)
-2
>>> round(-2.009,2)
-2.01
>>> round(2.675, 2) # note this rounds down
2.67
>>> max(4,-5,5,1,11)
11
>>> min(4,-5,5,1,11)
-5Many numeric functions are not built into the Python core and need to be included in our script if we want to use them. To include them at the tip of the script type:
import math
These next functions are found in the math module and need to be imported. To use these functions, prepend the function with the module name, i.e, math.ceil(15.5)
| math.function | Description |
|---|---|
math.ceil(x) |
return the smallest integer greater than or equal to x is returned |
math.floor(x) |
return the largest integer less than or equal to x. |
math.exp(x) |
The exponential of x: ex is returned |
math.log(x) |
the natural logarithm of x, for x > 0 is returned |
math.log10(x) |
The base-10 logarithm of x for x > 0 is returned |
math.modf(x) |
The fractional and integer parts of x are returned in a two-item tuple. |
math.pow(x, y) |
The value of x raised to the power y is returned |
math.sqrt(x) |
Return the square root of x for x >= 0 |
>>> import math
>>>
>>> math.ceil(2.3)
3
>>> math.ceil(2.9)
3
>>> math.ceil(-2.9)
-2
>>> math.floor(2.3)
2
>>> math.floor(2.9)
2
>>> math.floor(-2.9)
-3
>>> math.exp(2.3)
9.974182454814718
>>> math.exp(2.9)
18.17414536944306
>>> math.exp(-2.9)
0.05502322005640723
>>>
>>> math.log(2.3)
0.8329091229351039
>>> math.log(2.9)
1.0647107369924282
>>> math.log(-2.9)
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
ValueError: math domain error
>>>
>>> math.log10(2.3)
0.36172783601759284
>>> math.log10(2.9)
0.4623979978989561
>>>
>>> math.modf(2.3)
(0.2999999999999998, 2.0)
>>>
>>> math.pow(2.3,1)
2.3
>>> math.pow(2.3,2)
5.289999999999999
>>> math.pow(-2.3,2)
5.289999999999999
>>> math.pow(2.3,-2)
0.18903591682419663
>>>
>>> math.sqrt(25)
5.0
>>> math.sqrt(2.3)
1.51657508881031
>>> math.sqrt(2.9)
1.70293863659264Often times it is necessary to compare two numbers and find out if the first number is less than, equal to, or greater than the second.
The simple function cmp(x,y) is not available in Python 3.
Use this idiom instead:
cmp = (x>y)-(x<y)It returns three different values depending on x and y
-
if x<y -1 is returned
-
if x>y 1 is returned
-
x == y 0 is returned
In the next section, we will learn about strings, tuples and lists. These are all examples of python sequences. A string is a sequence of characters 'ACGTGA', a tuple (0.23, 9.74, -8.17, 3.24, 0.16) and a list ['dog', 'cat', 'bird'] are sequences of any kind of data. We'll see much more detail in a bit.
In Python a type of object gets operations that belong to that type. Sequences have sequence operations so strings can also use sequence operations. Strings also have their own specific operations.
You can ask what the length of any sequence is
>>>len('ACGTGA') # length of a string
6
>>>len( (0.23, 9.74, -8.17, 3.24, 0.16) ) # length of a tuple, needs two parentheses (( ))
5
>>>len(['dog', 'cat', 'bird']) # length of a list
3You can also use string-specific functions on strings, but not on lists and vice versa. We'll learn a lot more about this later on. rstrip() is a string method or function. You get an error if you try to use it on a list.
>>> 'ACGTGA'.rstrip('A')
'ACGTG'
>>> ['dog', 'cat', 'bird'].rstrip()
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
AttributeError: 'list' object has no attribute 'rstrip'How do you find out what functions work with an object? There's a handy function dir(). As an example what functions can you call on our string 'ACGTGA'?
>>> dir('ACGTGA')
['__add__', '__class__', '__contains__', '__delattr__', '__dir__', '__doc__', '__eq__', '__format__', '__ge__', '__getattribute__', '__getitem__', '__getnewargs__', '__gt__', '__hash__', '__init__', '__init_subclass__', '__iter__', '__le__', '__len__', '__lt__', '__mod__', '__mul__', '__ne__', '__new__', '__reduce__', '__reduce_ex__', '__repr__', '__rmod__', '__rmul__', '__setattr__', '__sizeof__', '__str__', '__subclasshook__', 'capitalize', 'casefold', 'center', 'count', 'encode', 'endswith', 'expandtabs', 'find', 'format', 'format_map', 'index', 'isalnum', 'isalpha', 'isdecimal', 'isdigit', 'isidentifier', 'islower', 'isnumeric', 'isprintable', 'isspace', 'istitle', 'isupper', 'join', 'ljust', 'lower', 'lstrip', 'maketrans', 'partition', 'replace', 'rfind', 'rindex', 'rjust', 'rpartition', 'rsplit', 'rstrip', 'split', 'splitlines', 'startswith', 'strip', 'swapcase', 'title', 'translate', 'upper', 'zfill']dir() will return all atributes of an object, among them its functions. Technically, functions belonging to a specific object are called methods.
You can call dir() on any object, most often, you'll use it in the interactive Python shell.
- A string is a series of characters starting and ending with single or double quotation marks.
- Strings are an example of a Python sequence. A sequence is defined as a positionally ordered set. This means each element in the set has a position, starting with zero, i.e. 0,1,2,3 and so on until you get to the end of the string.
- Single (')
- Double (")
- Triple (''' or """)
Notes about quotes:
- Single and double quotes are equivalent.
- A variable name inside quotes is just the string identifier, not the value stored inside the variable.
- Triple quotes are used before and after a string that spans multiple lines.
Use of quotation examples:
word = 'word'
sentence = "This is a sentence."
paragraph = """This is a paragraph. It is
made up of multiple lines and sentences. And goes
on and on.
"""We saw examples of print() earlier. Lets talk about it a bit more. print() is a function that takes one or more comma-separated arguments.
Let's use the print() function to print a string.
>>>print("ATG")
ATGLet's assign a string to a variable and print the variable.
>>>dna = 'ATG'
ATG
>>> print(dna)
ATGWhat happens if we put the variable in quotes?
>>>dna = 'ATG'
ATG
>>> print("dna")
dnaThe literal string 'dna' is printed to the screen, not the contents 'ATG'
Let's see what happens when we give print() two literal strings as arguments.
>>> print("ATG","GGTCTAC")
ATG GGTCTACWe get the two literal strings printed to the screen separated by a space
What if you do not want your strings separated by a space? Use the concatenation operator to concatenate the two strings before or within the print() function.
>>> print("ATG"+"GGTCTAC")
ATGGGTCTAC
>>> combined_string = "ATG"+"GGTCTAC"
ATGGGTCTAC
>>> print(combined_string)
ATGGGTCTACWe get the two strings printed to the screen without being separated by a space.
You can also use this
>>> print('ATG','GGTCTAC',sep='')
ATGGGTCTACNow, lets print a variable and a literal string.
>>>dna = 'ATG'
ATG
>>> print(dna,'GGTCTAC')
ATG GGTCTACWe get the value of the variable and the literal string printed to the screen separated by a space
How would we print the two without a space?
>>>dna = 'ATG'
ATG
>>> print(dna + 'GGTCTAC')
ATGGGTCTACSomething to think about: Values of variables are variable. Or in other words, they are mutable, changeable.
>>>dna = 'ATG'
ATG
>>> print(dna)
ATG
>>>dna = 'TTT'
TTT
>>> print(dna)
TTTThe new value of the variable 'dna' is printed to the screen when
dnais an argument for theprint()function.
Let's look at the typical errors you will encounter when you use the print() function.
What will happen if you forget to close your quotes?
>>> print("GGTCTAC)
File "<stdin>", line 1
print("GGTCTAC)
^
SyntaxError: EOL while scanning string literalWe get a 'SyntaxError' if the closing quote is not used
What will happen if you forget to enclose a string you want to print in quotes?
>>> print(GGTCTAC)
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
NameError: name 'GGTCTAC' is not defined
>>> GGTCTAC = 5 # define a variable
>>> print(GGTCTAC)
5We get a 'NameError' when the literal string is not enclosed in quotes because Python is looking for a variable with the name GGTCTAC
>>> print "boo"
File "<stdin>", line 1
print "boo"
^
SyntaxError: Missing parentheses in call to 'print'In python2, the command was print, but this changed to print() in python3, so don't forget the parentheses!
How would you include a new line, carriage return, or tab in your string?
| Escape Character | Description |
|---|---|
| \n | New line |
| \r | Carriage Return |
| \t | Tab |
Let's include some escape characters in our strings and print() functions.
>>> string_with_newline = 'this sting has a new line\nthis is the second line'
>>> print(string_with_newline)
this sting has a new line
this is the second lineWe printed a new line to the screen
print() adds spaces between arguments and a new line at the end for you. You can change these with sep= and end=. Here's an example:
print('one line', 'second line' , 'third line', sep='\n', end = '')
A neater way to do this is to express a multi-line string enclosed in triple quotes (""").
>>> print("""this string has a new line
... this is the second line""")
this string has a new line
this is the second lineLet's print a tab character (\t).
>>> line = "value1\tvalue2\tvalue3"
>>> print(line)
value1 value2 value3We get the three words separated by tab characters. A common format for data is to separate columns with tabs like this.
You can add a backslash before any character to force it to be printed as a literal. This is called 'escaping'. This is only really useful for printing literal quotes ' and "
>>> print('this is a \'word\'') # if you want to print a ' inside '...'
this is a 'word'
>>> print("this is a 'word'") # maybe clearer to print a ' inside "..."
this is a 'word'In both cases actual single quote character are printed to the screen
If you want every character in your string to remain exactly as it is, declare your string a raw string literal with 'r' before the first quote. This looks ugly, but it works.
>>> line = r"value1\tvalue2\tvalue3"
>>> print(line)
value1\tvalue2\tvalue3Our escape characters '\t' remain as we typed them, they are not converted to actual tab characters.
To concatenate strings use the concatenation operator '+'
>>> promoter= 'TATAAA'
>>> upstream = 'TAGCTA'
>>> downstream = 'ATCATAAT'
>>> dna = upstream + promoter + downstream
>>> print(dna)
TAGCTATATAAAATCATAATThe concatenation operator can be used to combine strings. The newly combined string can be stored in a variable.
What happens if you use + with numbers (these are integers or ints)?
>>> 4+3
7For strings, + concatenates; for integers, + adds.
You need to convert the numbers to strings before you can concatenate them
>>> str(4) + str(3)
'43'Use the len() function to calculate the length of a string. This function takes a sequence as an argument and returns an int
>>> print(dna)
TAGCTATATAAAATCATAAT
>>> len(dna)
20The length of the string, including spaces, is calculated and returned.
The value that len() returns can be stored in a variable.
>>> dna_length = len(dna)
>>> print(dna_length)
20You can mix strings and ints in print(), but not in concatenation.
>>> print("The lenth of the DNA sequence:" , dna , "is" , dna_length)
The lenth of the DNA sequence: TAGCTATATAAAATCATAAT is 20Changing the case of a string is a bit different than you might first expect. For example, to lowercase a string we need to use a method. A method is a function that is specific to an object. When we assign a string to a variable we are creating an instance of a string object. This object has a series of methods that will work on the data that is stored in the object. Recall that dir() will tell you all the methods that are available for an object. The lower() function is a string method.
Let's create a new string object.
dna = "ATGCTTG"Look familiar? It should!!! Creating a string object is what we have been doing all along!!! Jeez!!!
Now that we have a string object we can use string methods. The way you use a method is to put a '.' between the object and the method name.
>>> dna = "ATGCTTG"
>>> dna.lower()
'atgcttg'the lower() method returns the contents stored in the 'dna' variable in lowercase.
The contents of the 'dna' variable have not been changed. Strings are immutable. If you want to keep the lowercased version of the string, store it in a new variable.
>>> print(dna)
ATGCTTG
>>> dna_lowercase = dna.lower()
>>> print(dna)
ATGCTTG
>>> print(dna_lowercase)
atgcttgThe string method can be nested inside of other functions.
>>> dna = "ATGCTTG"
>>> print(dna.lower())
atgcttgThe contents of 'dna' are lowercased and passed to the
print()function.
If you try to use a string method on a object that is not a string you will get an error.
>>> nt_count = 6
>>> dna_lc = nt_count.lower()
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
AttributeError: 'int' object has no attribute 'lower'You get an AttributeError when you use a method on the an incorrect object type. We are told that the int object (an int is returned by
len()) does not have a function called lower.
Now let's uppercase a string.
>>> dna = 'attgct'
>>> dna.upper()
'ATTGCT'
>>> print(dna)
attgctThe contents of the variable 'dna' were returned in upper case. The contents of 'dna' were not altered.
count(str) returns the number of exact matches of str it found (as an int)
>>> dna = 'ATGCTGCATT'
>>> dna.count('T')
4The number of times 'T' is found is returned. The string stored in 'dna' is not altered.
replace(str1,str2) returns a new string with all matches of str1 in a string replaced with str2.
>>> dna = 'ATGCTGCATT'
>>> dna.replace('T','U')
'AUGCUGCAUU'
>>> print(dna)
ATGCTGCATT
>>> rna = dna.replace('T','U')
>>> print(rna)
AUGCUGCAUUAll occurrences of T are replaced by U. The new string is returned. The original string has not actually been altered. If you want to reuse the new string, store it in a variable.
Parts of a string can be located based on position and returned. This is because a string is a sequence. Coordinates start at 0. You add the coordinate in square brackets after the string's name.
This string 'ATTAAAGGGCCC' is made up of the following sequence of characters, and positions (starting at zero).
| Position/Index | Character |
|---|---|
| 0 | A |
| 1 | T |
| 2 | T |
| 3 | A |
| 4 | A |
| 5 | A |
| 6 | G |
| 7 | G |
| 8 | G |
| 9 | C |
| 10 | C |
| 11 | C |
Let's return the 4th, 5th, and 6th nucleotides. To do this, we need to count like a computer and start our string at 0 and return the 3rd, 4th, and 5th characters. This will be everything from 3 to 6. Python counts the gaps before each character in the string, starting at 0.
>>> dna = 'ATTAAAGGGCCC'
>>> sub_dna = dna[3:6]
>>> print(sub_dna)
AAAThe characters with indices 3, 4, 5 are returned. Or in other words, every character starting at index 3 and up to but not including, the index of 6 are returned.
Let's return the first 6 characters.
>>> dna = 'ATTAAAGGGCCC'
>>> sub_dna = dna[0:6]
>>> print(sub_dna)
ATTAAAEvery character starting at index 0 and up to but not including index 6 are returned. This is the same as dna[:6]
Let's return every character from index 6 to the end of the string.
>>> dna = 'ATTAAAGGGCCC'
>>> sub_dna = dna[6:]
>>> print(sub_dna)
GGGCCCWhen the second argument is left blank, every character from index 6 and greater is returned.
Let's return the last 3 characters.
>>> sub_dna = dna[-3:]
>>> print(sub_dna)
CCCWhen the second argument is left blank and the first argument is negative (-X), X characters from the end of the string are returned.
The positional index of an exact string in a larger string can be found and returned with the string method
find(). An exact string is given as an argument and the index of its first occurrence is returned. -1 is returned if it is not found.
>>> dna = 'ATTAAAGGGCCC'
>>> dna.find('T')
1
>>> dna.find('N')
-1The substring 'T' is found for the first time at index 1 in the string 'dna' so 1 is returned. The substring 'N' is not found, so -1 is returned.
Since these are methods, be sure to use in this format string.method().
| function | Description |
|---|---|
s.strip() |
returns a string with the whitespace removed from the start and end |
s.isalpha() |
tests if all the characters of the string are alphabetic characters. Returns True or False. |
s.isdigit() |
tests if all the characters of the string are numeric characters. Returns True or False. |
s.startswith('other_string') |
tests if the string starts with the string provided as an argument. Returns True or False. |
s.endswith('other_string') |
tests if the string ends with the string provided as an argument. Returns True or False. |
s.split('delim') |
splits the string on the given exact delimiter. Returns a list of substrings. If no argument is supplied, the string will be split on whitespace. |
s.join(list) |
opposite of split(). The elements of a list will be concatenated together using the string stored in 's' as a delimiter. |
Strings can be formated using the format() function. Pretty intuitive, but wait til you see the details! For example, if you want to include literal stings and variables in your print statement and do not want to concatenate or use multiple arguments in the print() function you can use string formatting.
>>> string = "This sequence: {} is {} nucleotides long and is found in {}."
>>> string.format(dna,dna_len,gene_name)
'This sequence: TGAACATCTAAAAGATGAAGTTT is 23 nucleotides long and is found in Brca1.'
>>> print(string) # string.format() does not alter string
This sequence: {} is {} nucleotides long and is found in {}.
>>> new_string = string.format(dna,dna_len,gene_name)
>>> print(new_string)
This sequence: TGAACATCTAAAAGATGAAGTTT is 23 nucleotides long and is found in Brca1.We put together the three variables and literal strings into a single string using the function format(). The original string is not altered, a new string is returned that incorporates the arguments. You can save the returned value in a new variable. Each {} is a placeholder for the strings that need to be inserted.
Something nice about format() is that you can print int and string variable types without converting first.
You can also directly call the format function on a string inside a print function. Here are two examples
>>> string = "This sequence: {} is {} nucleotides long and is found in {}."
>>> print(string.format(dna,dna_len,gene_name))
This sequence: TGAACATCTAAAAGATGAAGTTT is 23 nucleotides long and is found in Brca1.Or you use the format() function on a literal string:
>>> print( "This sequence: {} is {} nucleotides long and is found in {}.".format(dna,dna_len,gene_name))
This sequence: TGAACATCTAAAAGATGAAGTTT is 23 nucleotides long and is found in Brca1.There is no need to store the string in a variable.
So far, we have just used {} to show where to insert the value of a variable in a string. You can add special characters inside the {} to change the way the variable is formatted when it's inserted into the string.
You can number these, not necessarily in order.
>>> '{0}, {1}, {2}'.format('a', 'b', 'c')
'a, b, c'
>>> '{2}, {1}, {0}'.format('a', 'b', 'c')
'c, b, a'To change the spacing of strings and the way numbers are formatted, you add : and other special characters like this {:>5} to right-justify a string in a five-character field.
Lets right justify some numbers.
>>> print( "{:>5}".format(2) )
2
>>> print( "{:>5}".format(20) )
20
>>> print( "{:>5}".format(200) )
200How about padding with zeroes? This means the five-character field will be filled as needed with zeroes to the left of any numbers you want to display
>>> print( "{:>05}".format(2) )
00002
>>> print( "{:>05}".format(20) )
00020Use a < to indicate left-justification.
>>> print( "{:<5} genes".format(2) )
2 genes
>>> print( "{:<5} genes".format(20) )
20 genes
>>> print( "{:<5} genes".format(200) )
200 genesCenter aligning is done with ^ instead of > or <. You can also pad with characters other than 0. Here let's try _ or underscore as in :_^.
>>> print( "{:_^10}".format(2) )
____2_____
>>> print( "{:_^10}".format(20) )
____20____
>>> print( "{:_^10}".format(200) )
___200____Text can be centered by using ':^10'. 10 of course is your column width. The '^' indicates center justification. In our example an underscore is used to replace the empty spaces to make things easier to see.
Here are some of the ALIGNMENT options:
| Option | Meaning | |
|---|---|---|
| '<' | Forces the field to be left-aligned within the available space (this is the default for most objects). | |
| '>' | Forces the field to be right-aligned within the available space (this is the default for numbers). | |
| '=' | Forces the padding to be placed after the sign (if any) but before the digits. This is used for printing fields in the form ‘+000000120’. This alignment option is only valid for numeric types. | |
| '^' | Forces the field to be centered within the available space. |
Here's an example
{ : x < 10 s}fill with
x
left justify<
10a field of ten characterssa string
Common Types
| type | description |
|---|---|
| b | convert to binary |
| d | decimal integer |
| e | exponent, default precision is 6, uses e |
| E | exponent, uses E |
| f | floating point, default precision 6 (also F) |
| g | general number, float for values close to 0, exponent for others; also G |
| s | string, default type (see example above) |
| x | convert to hexadecimal, also X |
| % | converts to % by multiplying by 100 |
So much can be done with the format() function. Here is one last example, but not the last functionality of this function. Let truncate a long floating point number. The default is 6 decimal places. Note that the function rounds to the nearest decimal place, but not always exactly the way you expect because of the way computers represent decimals with 1s and 0s.
'{:f}'.format(3.141592653589793)
'3.141593'
>>> '{:.4f}'.format(3.141592653589793)
'3.1416'Lists are data types that store a collection of data.
- Lists are used to store an ordered, indexed collection of data.
- Values are separated by commas
- Values are enclosed in square brackets '[]'
- Lists can grow and shrink
- Values are mutable
- Tuples are used to store an ordered, indexed collection of data.
- Values are separated by commas
- Values are enclosed in square brackets '()'
- Tuples can NOT grow or shrink
- Values are immutable
To retrieve a single value in a list use the value's index in this format list[index]. This will return the value at the specified index, starting with 0.
Here is a list:
>>> codons = [ 'atg' , 'aaa' , 'agg' ]There are 3 values with the indices of 0, 1, 2
| Index | Value |
|---|---|
| 0 | atg |
| 1 | aaa |
| 2 | agg |
Let's access the 0th value, this is the element in the list with index 0. You'll need an index number (0) inside square brackets like this [0] . This goes after the name of the list (codons)
>>> codons = [ 'atg' , 'aaa' , 'agg' ]
>>> codons[0]
'atg'The value can be saved for later use by storing in a variable.
>>> codons = [ 'atg' , 'aaa' , 'agg' ]
>>> first_codon = codons[0]
>>> print(first_codon)
atgEach value can be saved in a new variable to use later.
The values can be retrieved and used directly.
>>> codons = [ 'atg' , 'aaa' , 'agg' ]
>>> print(codons[0])
atg
>>> print(codons[1])
aaa
>>> print(codons[2])
aggThe 3 values are independently accesses and immediately printed. They are not stored in a variable.
If you want to access the values starting at the end of the list, use negative indices.
>>> codons = [ 'atg' , 'aaa' , 'agg' ]
>>> print(codons[-1])
agg
>>> print(codons[-2])
aaaUsing a negative index will return the values from the end of the list. For example, -1 is the index of the last value 'agg'. This value also has an index of 2.
Individual values can be changed using the value's index and the assignment operator.
>>> print(codons)
['atg', 'aaa', 'agg']
>>> codons[2] = 'cgc'
>>> print(codons)
['atg', 'aaa', 'cgc']What about trying to assign a value to an index that does not exist?
>>> codons[5] = 'aac'
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
IndexError: list assignment index out of rangecodon[5] does not exist, and when we try to assign a value to this index we get an IndexError. If you want to add new elements to the end of a list use
codons.append('taa')orcodons.extend(list). See below for more details.
This works in exactly the same way with lists as it does with strings. This is because both are sequences, or ordered collections of data with positional information. Remember Python counts the divisions between the elements, starting with 0.
| Index | Value |
|---|---|
| 0 | atg |
| 1 | aaa |
| 2 | agg |
| 3 | aac |
| 4 | cgc |
| 5 | acg |
>>> codons = [ 'atg' , 'aaa' , 'agg' , 'aac' , 'cgc' , 'acg']
>>> print (codons[1:3])
['aaa', 'agg']
>>> print (codons[3:])
['aac', 'cgc', 'acg']
>>> print (codons[:3])
['atg', 'aaa', 'agg']
>>> print (codons[0:3])
['atg', 'aaa', 'agg']
codons[1:3]returns every value starting with the value of codons[1] up to but not including codons[3]codons[3:]returns every value starting with the value of codons[3] and every value after.codons[:3]returns every value up to but not including codons[3]codons[0:3]is the same ascodons[:3]
| Operator | Description | Example |
|---|---|---|
+ |
Concatenation | [10, 20, 30] + [40, 50, 60] returns [10, 20, 30, 40, 50, 60] |
* |
Repetition | ['atg'] * 4 returns ['atg','atg','atg','atg'] |
in |
Membership | 20 in [10, 20, 30] returns True |
| Functions | Description | Example |
|---|---|---|
len(list) |
returns the length or the number of values in list | len([1,2,3]) returns 3 |
max(list) |
returns the value with the highest ASCII value (=latest in ASCII alphabet) | max(['a','A','z']) returns 'z' |
min(list) |
returns the value with the lowest ASCII value (=earliest in ASCII alphabet) | min(['a','A','z']) returns 'A' |
list(seq) |
converts a tuple into a list | list(('a','A','z')) returns ['a', 'A', 'z'] |
sorted(list, key=None, reverse=False) |
returns a sorted list based on the key provided | sorted(['a','A','z']) returns ['A', 'a', 'z'] |
str.lower() makes all the elements lowercase before sorting |
sorted(['a','A','z'],key=str.lower) returns ['a', 'A', 'z'] |
Remember methods are used in the following format list.method().
For these examples use: nums = [1,2,3] and codons = [ 'atg' , 'aaa' , 'agg' ]
| Method | Description | Example |
|---|---|---|
list.append(obj) |
appends an object to the end of a list | nums.append(9) ; print(list) ; returns [1,2,3,9] |
list.count(obj) |
counts the occurrences of an object in a list | nums.count(2) returns 1 |
list.index(obj) |
returns the lowest index where the given object is found | nums.index(2) returns 1 |
list.pop() |
removes and returns the last value in the list. The list is now one element shorter | nums.pop() returns 3 |
list.insert(index, obj) |
inserts a value at the given index. Remember to think about the divisions between the elements | list.insert(0,100) ; print(list) returns [100, 1, 2, 3] |
list.extend(new_list) |
appends new_list to the end of list |
list.extend([7, 8]) ; print(list) returns [1, 2, 3, 7,8] |
list.pop(index) |
removes and returns the value of the index argument. The list is now 1 value shorter | nums.pop(0) returns 1 |
list.remove(obj) |
finds the lowest index of the given object and removes it from the list. The list is now one element shorter | codons.remove('aaa') ; print(codons) returns [ 'atg' , 'agg' ] |
list.reverse() |
reverses the order of the list | nums.reverse() ; print(nums) returns [3,2,1] |
list.copy() |
Returns a shallow copy of list. Shallow vs Deep only matters in multidimensional data structures. | |
list.sort([func]) |
sorts a list using the provided function. Does not return a list. The list has been changed. Advanced list sort will be covered once writing your own functions has been discussed. | codons.sort() ; print(codons) returns ['aaa', 'agg', 'atg'] |
Be careful how you make a copy of your list
>>> my_list=['a', 'one', 'two']
>>> copy_list=my_list
>>> copy_list.append('1')
>>> print(my_list)
['a', 'one', 'two', '1']
>>> print(copy_list)
['a', 'one', 'two', '1']Not what you expected?! Both lists have changed because we only copied a pointer to the original list when we wrote
copy_list=my_list.
Let's copy the list using the copy() method.
>>> my_list=['a', 'one', 'two']
>>> copy_list=my_list.copy()
>>> copy_list.append('1')
>>> print(my_list)
['a', 'one', 'two']There we go, we get what we expect this time!
Now that you have seen the append() function we can go over how to build a list one value at a time.
>>> words = []
>>> print(words)
[]
>>> words.append('one')
>>> words.append('two')
>>> print(words)
['one', 'two']We start with a an empty list called 'words'. We use
append()to add the value 'one' then to add the value 'two'. We end up with a list with two values. You can add a whole list to another list withwords.extend(['three','four','five'])
All of the coding that we have gone over so far has been executed line by line. Sometimes there are blocks of code that we want to execute more than once. Loops let us do this.
There are two loop types:
- while loop
- for loop
The while loop will continue to execute a block of code as long as the test expression evaluates to True.
while expression:
statement1
statement2
more_statements
rest_of_code_goes_here
The condition is the expression. The while loop block of code is the collection of indented statements following the expression.
Code:
#!/usr/bin/env python3
count = 0
while count < 5:
print("count:" , count)
count+=1
print("Done") Output:
$ python while.py
count: 0
count: 1
count: 2
count: 3
count: 4
Done
The while condition was true 5 times and the while block of code was executed 5 times.
- count is equal to 0 when we begin
- 0 is less than 5 so we execute the while block
- count is printed
- count is incremented (count = count + 1)
- count is now equal to 1.
- 1 is less than 5 so we execute the while block for the 2nd time.
- this continues until count is 5.
- 5 is not less than 5 so we exit the while block
- The first line following the while statement is executed, "Done" is printed
An infinite loop occurs when a while condition is always true. Here is an example of an infinite loop.
#!/usr/bin/env python3
count = 0
while count < 5:
print("count:" , count)
print("Done") Output:
$ python infinite.py
count: 0
count: 0
count: 0
count: 0
count: 0
count: 0
count: 0
count: 0
...
...
What caused the expression to always be
True? The statement that increments the count is missing, so it will always be smaller than 5. To stop the code from forever printing use ctrl+c.
An Else statment can be used with a while statement. It behaves in the same way as and Else with an If statement. When the while statement is false, the else block is executed ONE TIME.
#!/usr/bin/env python3
count = 0
while count < 5:
print("count:" , count)
count+=1
else:
print("count:", count, "is now not less than 5")
print("Done")Output:
$ python while_else.py
count: 0
count: 1
count: 2
count: 3
count: 4
count: 5 is now not less than 5
Done
The while loop was executed five times like before. Now when count is equal to 5 and therefore not less than 5, the else block is executed. Finally the lines of code outside the while/else are executed.
A for loop is a loop that executes the for block of code for every member of a sequence, for example the elements of a list or the letters in a string.
for iterating_variable in sequence:
statement(s)An example of a sequence is a list. Let's use a for loop with a list of words.
Code:
#!/usr/bin/env python3
words = ['zero','one','two','three','four']
for word in words:
print(word)Notice how I have named my variables, the list is plural and the iterating variable is singular
Output:
python3 list_words.py
zero
one
two
three
four
This is next example is using a for loop to iterating over a string. Remember a string is a sequence like a list. Each character has a position. Look back at "Extracting a Substring, or Slicing" in the Strings section to see other ways that strings can be treated like lists.
Code:
#!/usr/bin/env python3
dna = 'GTACCTTGATTTCGTATTCTGAGAGGCTGCTGCTTAGCGGTAGCCCCTTGGTTTCCGTGGCAACGGAAAA'
for nt in dna:
print(nt)Output:
$ python3 for_string.py
G
T
A
C
C
T
T
...
...
This is an easy way to access each character in a string. It is especially nice for DNA sequences.
Another example of iterating over a list of variables, this time numbers.
Code:
#!/usr/bin/env python3
numbers = [0,1,2,3,4]
for num in numbers:
print(num)Output:
$ python3 list_numbers.py
0
1
2
3
4
Python has a function called range() that will return numbers that can be converted to a list.
>>> range(5)
range(0, 5)
>>> list(range(5))
[0, 1, 2, 3, 4]The function range() can be used in conjunction with a for loop to iterate over a range of numbers. Range also starts at 0 and thinks about the gaps between the numbers.
Code:
#!/usr/bin/env python3
for num in range(5):
print(num)Output:
$ python list_range.py
0
1
2
3
4
As you can see this is the same output as using the list
numbers = [0, 1, 2, 3, 4]And this has the same functionality as a while loop with a condition ofcount = 0;count < 5.
This is the equivalent while loop
Code:
count = 0
while count < 5:
print(count)
count+=1Output:
0
1
2
3
4
An else statement can be used with a for loop as well. The else block of code will be executed when the for loop exits normally.
Code:
#!/usr/bin/env python3
for num in range(5):
print(num)
else:
print("Completed for loop")Output:
$ python3 list_range_else.py
0
1
2
3
4
Completed for loop
Loops control statements allow for altering the normal flow of execution.
| Control Statement | Description |
|---|---|
break |
A loop is terminated when a break statement is executed. All the lines of code after the break, but within the loop block are not executed. No more iteration of the loop are preformed |
continue |
A single iteration of a loop is terminated when a continue statement is executed. The next iteration will proceed normally. |
Code:
#!/usr/bin/env python3
count = 0
while count < 5:
print("count:" , count)
count+=1
if count == 3:
break
print("Done")Output:
$ python break.py
count: 0
count: 1
count: 2
Done
when the count is equal to 3, the execution of the while loop is terminated, even though the initial condition (count < 5) is still True.
Code:
#!/usr/bin/env python3
count = 0
while count < 5:
print("count:" , count)
count+=1
if count == 3:
continue
print("line after our continue")
print("Done")Output:
$ python continue.py
count: 0
line after our continue
count: 1
line after our continue
count: 2
count: 3
line after our continue
count: 4
line after our continue
Done
When the count is equal to 3 the continue is executed. This causes all the lines within the loop block to be skipped. "line after our continue" is not printed when count is equal to 3. The next loop is executed normally.
An iterable is any data type that is can be iterated over, or can be used in iteration. An iterable can be made into an iterator with the iter() function. This means you can use the next() function.
>>> codons = [ 'atg' , 'aaa' , 'agg' ]
>>> codons_iterator=iter(codons)
>>> next(codons_iterator)
'atg'
>>> next(codons_iterator)
'aaa'
>>> next(codons_iterator)
'agg'
>>> next(codons_iterator)
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
StopIterationAn iterator allows you to get the next element in the iterator until there are no more elements. If you want to go through each element again, you will need to redefine the iterator.
Example of using an iterator in a for loop:
codons = [ 'atg' , 'aaa' , 'agg' ]
>>> codons_it = iter(codons)
>>> for codon in codons_it :
... print( codon )
...
atg
aaa
aggThis is nice if you have a large large large list that you don't want to keep in memory. An iterator allows you to go through each element but not keep the entire list in memory. Without iterators the entire list is in memory.
List comprehension is a way to make a list without typing out each element. There are many many ways to use list comprehension to generate lists. Some are quite complex, yet useful.
Here is an simple example:
>>> dna_list = ['TAGC', 'ACGTATGC', 'ATG', 'ACGGCTAG']
>>> lengths = [len(dna) for dna in dna_list]
>>> lengths
[4, 8, 3, 8]This is how you could do the same with a for loop:
>>> lengths = []
>>> dna_list = ['TAGC', 'ACGTATGC', 'ATG', 'ACGGCTAG']
>>> for dna in dna_list:
... lengths.append(len(dna))
...
>>> lengths
[4, 8, 3, 8]Using conditions:
This will only return the length of an element that starts with 'A':
>>> dna_list = ['TAGC', 'ACGTATGC', 'ATG', 'ACGGCTAG']
>>> lengths = [len(dna) for dna in dna_list if dna.startswith('A')]
>>> lengths
[8, 3, 8]This generates the following list: [8, 3, 8]
Here is an example of using mathematical operators to generate a list:
>>> two_power_list = [2 ** x for x in range(10)]
>>> two_power_list
[1, 2, 4, 8, 16, 32, 64, 128, 256, 512]This creates a list of the of the product of [2^0 , 2^1, 2^2, 2^3, 2^4, 2^5, 2^6, 2^7, 2^8, 2^9 ]
Dictionaries are another iterable, like a string and list. Unlike strings and lists, dictionaries are not a sequence, or in other words, they are unordered and the position is not important.
Dictionaries are a collection of key/value pairs. In Python, each key is separated from its value by a colon (:), the items are separated by commas, and the whole thing is enclosed in curly braces. An empty dictionary without any items is written with just two curly braces, like this: {}
Each key in a dictionary is unique, while values may not be. The values of a dictionary can be of any type, but the keys must be of an immutable data type such as strings, numbers, or tuples.
Data that is appropriate for dictionaries are two pieces of information that naturally go together, like gene name and sequence.
| Key | Value |
|---|---|
| TP53 | GATGGGATTGGGGTTTTCCCCTCCCATGTGCTCAAGACTGGCGCTAAAAGTTTTGAGCTTCTCAAAAGTC |
| BRCA1 | GTACCTTGATTTCGTATTCTGAGAGGCTGCTGCTTAGCGGTAGCCCCTTGGTTTCCGTGGCAACGGAAAA |
genes = { 'TP53' : 'GATGGGATTGGGGTTTTCCCCTCCCATGTGCTCAAGACTGGCGCTAAAAGTTTTGAGCTTCTCAAAAGTC' , 'BRCA1' : 'GTACCTTGATTTCGTATTCTGAGAGGCTGCTGCTTAGCGGTAGCCCCTTGGTTTCCGTGGCAACGGAAAA' }Breaking up the key/value pairs over multiple lines make them easier to read.
genes = {
'TP53' : 'GATGGGATTGGGGTTTTCCCCTCCCATGTGCTCAAGACTGGCGCTAAAAGTTTTGAGCTTCTCAAAAGTC' ,
'BRCA1' : 'GTACCTTGATTTCGTATTCTGAGAGGCTGCTGCTTAGCGGTAGCCCCTTGGTTTCCGTGGCAACGGAAAA'
}To retrieve a single value in a dictionary use the value's key in this format dict[key]. This will return the value at the specified key.
>>> genes = { 'TP53' : 'GATGGGATTGGGGTTTTCCCCTCCCATGTGCTCAAGACTGGCGCTAAAAGTTTTGAGCTTCTCAAAAGTC' , 'BRCA1' : 'GTACCTTGATTTCGTATTCTGAGAGGCTGCTGCTTAGCGGTAGCCCCTTGGTTTCCGTGGCAACGGAAAA' }
>>>
>>> genes['TP53']
GATGGGATTGGGGTTTTCCCCTCCCATGTGCTCAAGACTGGCGCTAAAAGTTTTGAGCTTCTCAAAAGTCThe sequence of the gene TP53 is stored as a value of the key 'TP53'. We can assess the sequence by using the key in this format dict[key]
The value can be accessed and passed directly to a function or stored in a variable.
>>> print(genes['TP53'])
GATGGGATTGGGGTTTTCCCCTCCCATGTGCTCAAGACTGGCGCTAAAAGTTTTGAGCTTCTCAAAAGTC
>>>
>>> seq = genes['TP53'];
>>> print(seq)
GATGGGATTGGGGTTTTCCCCTCCCATGTGCTCAAGACTGGCGCTAAAAGTTTTGAGCTTCTCAAAAGTCIndividual values can be changed by using the key and the assignment operator.
>>> genes = { 'TP53' : 'GATGGGATTGGGGTTTTCCCCTCCCATGTGCTCAAGACTGGCGCTAAAAGTTTTGAGCTTCTCAAAAGTC' , 'BRCA1' : 'GTACCTTGATTTCGTATTCTGAGAGGCTGCTGCTTAGCGGTAGCCCCTTGGTTTCCGTGGCAACGGAAAA' }
>>>
>>> print(genes)
{'BRCA1': 'GTACCTTGATTTCGTATTCTGAGAGGCTGCTGCTTAGCGGTAGCCCCTTGGTTTCCGTGGCAACGGAAAA', 'TP53': 'GATGGGATTGGGGTTTTCCCCTCCCATGTGCTCAAGACTGGCGCTAAAAGTTTTGAGCTTCTCAAAAGTC'}
>>>
>>> genes['TP53'] = 'atg'
>>>
>>> print(genes)
{'BRCA1': 'GTACCTTGATTTCGTATTCTGAGAGGCTGCTGCTTAGCGGTAGCCCCTTGGTTTCCGTGGCAACGGAAAA', 'TP53': 'atg'}The contents of the dictionary have changed.
Other assignment operators can also be used to change a value of a dictionary key.
>>> genes = { 'TP53' : 'GATGGGATTGGGGTTTTCCCCTCCCATGTGCTCAAGACTGGCGCTAAAAGTTTTGAGCTTCTCAAAAGTC' , 'BRCA1' : 'GTACCTTGATTTCGTATTCTGAGAGGCTGCTGCTTAGCGGTAGCCCCTTGGTTTCCGTGGCAACGGAAAA' }
>>>
>>> genes['TP53'] += 'TAGAGCCACCGTCCAGGGAGCAGGTAGCTGCTGGGCTCCGGGGACACTTTGCGTTCGGGCTGGGAGCGTG'
>>>
>>> print(genes)
{'BRCA1': 'GTACCTTGATTTCGTATTCTGAGAGGCTGCTGCTTAGCGGTAGCCCCTTGGTTTCCGTGGCAACGGAAAA', 'TP53': 'GATGGGATTGGGGTTTTCCCCTCCCATGTGCTCAAGACTGGCGCTAAAAGTTTTGAGCTTCTCAAAAGTCTAGAGCCACCGTCCAGGGAGCAGGTAGCTGCTGGGCTCCGGGGACACTTTGCGTTCGGGCTGGGAGCGTG'}Here we have used the '+=' concatenation assignment operator. This is equivalent to genes['TP53'] = genes['TP53'] + 'TAGAGCCACCGTCCAGGGAGCAGGTAGCTGCTGGGCTCCGGGGACACTTTGCGTTCGGGCTGGGAGCGTG'.
Since a dictionary is a sequence we can iterate through its contents.
A for loop can be used to retrieve each key of a dictionary one a time:
>>> for gene in genes:
... print(gene)
...
TP53
BRCA1Once you have the key you can retrieve the value:
>>> for gene in genes:
... seq = genes[gene]
... print(gene, seq[0:10])
...
TP53 GATGGGATTG
BRCA1 GTACCTTGATBuilding a dictionary one key/value at a time is akin to what we just saw when we change a key's value. Normally you won't do this. We'll talk about ways to build a dictionary from a file in a later lecture.
>>> genes = {}
>>> print(genes)
{}
>>> genes['Brca1'] = 'GTACCTTGATTTCGTATTCTGAGAGGCTGCTGCTTAGCGGTAGCCCCTTGGTTTCCGTGGCAACGGAAAA'
>>> genes['TP53'] = 'GATGGGATTGGGGTTTTCCCCTCCCATGTGCTCAAGACTGGCGCTAAAAGTTTTGAGCTTCTCAAAAGTC'
>>> print(genes)
{'Brca1': 'GTACCTTGATTTCGTATTCTGAGAGGCTGCTGCTTAGCGGTAGCCCCTTGGTTTCCGTGGCAACGGAAAA', 'TP53': 'GATGGGATTGGGGTTTTCCCCTCCCATGTGCTCAAGACTGGCGCTAAAAGTTTTGAGCTTCTCAAAAGTC'}We start by creating an empty dictionary. Then we add each key/value pair using the same syntax as when we change a value.
dict[key] = new_value
Python generates an error (NameError) if you try to access a key that does not exist.
>>> print(genes['HDAC'])
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
NameError: name 'HDAC' is not defined| Operator | Description |
|---|---|
in |
key in dict returns True if the key exists in the dictionary |
not in |
key not in dict returns True if the key does not exist in the dictionary |
Because Python generates a NameError if you try to use a key that doesn't exist in the dictionary, you need to check whether a key exists before trying to use it.
The best way to check whether a key exists is to use in
>>> gene = 'TP53'
>>> if gene in genes:
... print('found')
...
found
>>>
>>> if gene in genes:
... print(genes[gene])
...
GATGGGATTGGGGTTTTCCCCTCCCATGTGCTCAAGACTGGCGCTAAAAGTTTTGAGCTTCTCAAAAGTC
>>>
If you want to print the contents of a dictionary, you probably want to sort the keys then iterate over the keys with a for loop. Why do you want to sort the keys?
for gene_key in sorted(genes):
print(gene_key, '=>' , genes[gene_key])This will print the same order of keys every time you run your script.
| Function | Description |
|---|---|
len(dict) |
returns the total number of key/value pairs |
str(dict) |
returns a string representation of the dictionary |
type(variable) |
Returns the type or class of the variable passed to the function. If the variable is dictionary, then it would return a dictionary type. |
These functions work on several other data types too!
| Method | Description |
|---|---|
dict.clear() |
Removes all elements of dictionary dict |
dict.copy() |
Returns a shallow copy of dictionary dict. Shallow vs. deep copying only matters in multidimensional data structures. |
dict.fromkeys(seq,value) |
Create a new dictionary with keys from seq (Python sequence type) and values set to value. |
dict.items() |
Returns a list of (key, value) tuple pairs |
dict.keys() |
Returns list of keys |
dict.get(key, default = None) |
get value from dict[key], use default if not not present |
dict.setdefault(key, default = None) |
Similar to get(), but will set dict[key] = default if key is not already in dict |
dict.update(dict2) |
Adds dictionary dict2's key-values pairs to dict |
dict.values() |
Returns list of dictionary dict's values |
A set is another Python data type. It is essentially a dictionary with keys but no values.
- A set is unordered
- A set is a collection of data with no duplicate elements.
- Common uses include looking for differences and eliminating duplicates in data sets.
Curly braces {} or the set() function can be used to create sets.
Note: to create an empty set you have to use
set(), not{}the latter creates an empty dictionary.
>>> basket = {'apple', 'orange', 'apple', 'pear', 'orange', 'banana'}
>>> print(basket)
{'orange', 'banana', 'pear', 'apple'}Look, duplicates have been removed
Test to see if an value is in the set
>>> 'orange' in basket
True
>>> 'crabgrass' in basket
FalseThe in operator works the same with sets as it does with lists and dictionaries
Union, intersection, difference and symmetric difference can be done with sets
>>> a = set('abracadabra')
>>> b = set('alacazam')
>>> a
{'a', 'r', 'b', 'c', 'd'}Sets contain unique elements, therefore, even if duplicate elements are provided they will be removed.
Difference
The difference between two sets are the elements that are unique to the set to the left of the - operator, with duplicates removed.
>>> a = set('abracadabra')
>>> b = set('alacazam')
>>> a - b
{'r', 'd', 'b'}This results the letters that are in a but not in b
Union
The union between two sets is a sequence of the all the elements of the first and second sets combined, with duplicates removed.
>>> a = set('abracadabra')
>>> b = set('alacazam')
>>> a | b
{'a', 'c', 'r', 'd', 'b', 'm', 'z', 'l'}This returns letters that are in a or b both
Intersection
The intersection between two sets is a sequence of the elements which are in both sets, with duplicates removed.
>>> a = set('abracadabra')
>>> b = set('alacazam')
>>> a & b
{'a', 'c'}This returns letters that are in both a and b
Symmetric Difference
The symmetric difference is the elements that are only in the first set plus the elements that are only in the second set, with duplicates removed.
>>> a = set('abracadabra')
>>> b = set('alacazam')
>>> a ^ b
{'r', 'd', 'b', 'm', 'z', 'l'}This returns the letters that are in a or b but not in both (also known as exclusive or)
| Function | Description |
|---|---|
all() |
returns True if all elements of the set are true (or if the set is empty). |
any() |
returns True if any element of the set is true. If the set is empty, return False. |
enumerate() |
returns an enumerate object. It contains the index and value of all the items of set as a pair. |
len() |
returns the number of items in the set. |
max() |
returns the largest item in the set. |
min() |
returns the smallest item in the set. |
sorted() |
returns a new sorted list from elements in the set (does not alter the original set). |
sum() |
returns the sum of all elements in the set. |
| Method | Description |
|---|---|
set.add(new) |
adds a new element |
set.clear() |
remove all elements |
set.copy() |
returns a shallow copy of a set |
set.difference(set2) |
returns the difference of set and set2 |
set.difference_update(set2) |
removes all elements of another set from this set |
set.discard(element) |
removes an element from set if it is found in set. (Do nothing if the element is not in set) |
set.intersection(sets) |
return the intersection of set and the other provided sets |
set.intersection_update(sets) |
updates set with the intersection of set and the other provided sets |
set.isdisjoint(set2) |
returns True if set and set2 have no intersection |
set.issubset(set2) |
returns True if set2 contains set |
set.issuperset(set2) |
returns True if set contains set2 |
set.pop() |
removes and returns an arbitrary element of set. |
set.remove(element) |
removes element from a set. |
set.symmetric_difference(set2) |
returns the symmetric difference of set and set2 |
set.symmetric_difference_update(set2) |
updates set with the symmetric difference of set and set2 |
set.union(sets) |
returns the union of set and the other provided sets |
set.update(set2) |
update set with the union of set and set2 |
I/O stands for input/output. The in and out refer to getting data into and out of your script. It might be a little surprising at first, but writing to the screen, reading from the keyboard, reading from a file, and writing to a file are all examples of I/O.
You should be well versed in writing to the screen. We have been using the print() function to do this.
>>> print ("Hello, PFB2017!")
Hello, PFB2017!Remember this example from one of our first lessons?
This is something new. There is a function which prints a message to the screen and waits for input from the keyboard. This input can be stored in a variable. It always starts as a string. Convert to an int or float if you want a number.
>>> user_input = input("Type Something Now: ")
Type Something Now: Hi
>>> print(user_input)
Hi
>>> type(user_input)
<class 'str'>Most of the data we will be dealing with will be contained in files.
The first thing to do with a file is open it. We can do this with the open() function. The open() function takes the file name and access mode as arguments and returns a file object.
The most common access modes are read (r) and write (w).
>>> file_object = open("seq.nt.fa","r")'file_object' is a name of a variable. This can be anything, but make it a helpful name that describes what kind of file you are opening.
Now that we have opened a file and created a file object we can do things with it, like read it. Lets read all the contents at once.
Let's go to the command line and cat the contents of the file to see what's in it first
$ cat seq.nt.fa
ACAAAATACGTTTTGTAAATGTTGTGCTGTTAACACTGCAAATAAACTTGGTAGCAAACACTTCCAAAAG
ACCGGTTTCCAAAGACAGTCTTCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGG
$ Note the new lines. Now, lets print the contents to the screen with Python. We will use read() to read the entire contents of the file into a variable.
>>> file = open("seq.nt.fa","r")
>>> contents = file.read()
seq.nt.fa
>>> print(contents) # note newline characters are part of the file!
ACAAAATACGTTTTGTAAATGTTGTGCTGTTAACACTGCAAATAAACTTGGTAGCAAACACTTCCAAAAG
ACCGGTTTCCAAAGACAGTCTTCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGG
>>> file.close()The complete contents can be retrieved with the
read()method. Notice the newlines are maintained whencontentsis printed to the screen.print()adds another new line when it is finished printing. It is good practice to close your file. Use theclose()method.
Here's another way to read data in from a file. A for loop can be used to iterate through the file one line at a time.
#!/usr/bin/env python3
file = open("seq.nt.fa","r")
for line in file: # Python magic: reads in a line from file
print(line)Output:
$ python3 file_for.py
ACAAAATACGTTTTGTAAATGTTGTGCTGTTAACACTGCAAATAAACTTGGTAGCAAACACTTCCAAAAG
ACCGGTTTCCAAAGACAGTCTTCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGG
Notice the blank line at after each line we print.
print()adds a newline and we have a newline at the end of each line in our file. Userstrip()method to remove the newline from each line.
Let's use rstrip() method to remove the newline from our file input.
$ cat file_for_rstrip.py
#!/usr/bin/env python3
file_object = open("seq.nt.fa","r")
for line in file_object:
line = line.rstrip()
print(line)
rstrip()without any parameters returns a string with whitespace removed from the end.
Output:
$ python3 file_for_rstrip.py
ACAAAATACGTTTTGTAAATGTTGTGCTGTTAACACTGCAAATAAACTTGGTAGCAAACACTTCCAAAAG
ACCGGTTTCCAAAGACAGTCTTCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGG
Where do the newlines in the above output come from?
####Opening a file with with open() as fh:
Many people add this because it closes the file for you automatically. Good programming practice. Your code will clean up as it runs. For more advanced coding, with ... as ... saves limited resources like filehandles and database connections. For now, we just need to know that the with ... as ...: does the same as fh = open(...) ... fh.close(). So here's what the adapted code looks like
#!/usr/bin/env python3
with open("seq.nt.fa","r") as file_object: #cleans up after exiting with block
for line in file_object:
line = line.rstrip()
print(line)
#file gets closed for you here.Writing to a file is nothing more than opening a file for writing then using the write() method.
The write() method is like the print() function. The biggest difference is that it writes to your file object instead of the screen. Unlike print() it does not add a newline by default. write() takes a single string argument.
Let's write a few lines to a file named "writing.txt".
#!/usr/bin/env python3
fo = open("writing.txt" , "w")
fo.write("One line.\n")
fo.write("2nd line.\n")
fo.write("3rd line" + " has extra text\n")
some_var = 5
fo.write("4th line has " + str(some_var) + " words\n")
fo.close()
print("Wrote to file 'writing.txt'") # it's nice to tell the user you wrote a fileOutput:
$ python3 file_write.py
Wrote to file 'writing.txt'
$ cat writing.txt
One line.
2nd line.
3rd line has extra text
4th line has 5 words
Now, let's get crazy! Lets read from one file a line at a time. Do something to each line and write the results to a new file.
#!/usr/bin/env python3
seq_read = open("seq.nt.fa","r")
seq_write = open("nt.counts.txt","w")
total_nts = 0
for line in seq_read:
line = line.rstrip()
nt_count = len(line)
total_nts += nt_count
seq_write.write(str(nt_count) + "\n")
seq_write.write("Total: " + str(total_nts) +"\n")
seq_read.close()
seq_write.close()
print("Wrote 'nt.counts.txt'")Output:
$ python file_read_write.py
$ cat nt.counts.txt
71
71
Total: 142
The file we are reading from is named, "seq.nt.fa"
The file we are writing to is named, "nt.counts.txt"
We read each line, calculate the length of each line and print the length
We also create a variable to keep track of the total nt count
At the end, we print out the total count of nts
Finally we close each of the files
This is a very common task. It will use a loop, file I/O and a dictionary.
Assume we have a file called "sequence_data.txt" that contains tab-delimited gene names and sequences that looks something like this
TP53 GATGGGATTGGGGTTTTCCCCTCCCATGTGCTCAAGACTGGCGCTAAAAGTTTTGAGCTTCTCAAAAGTC
BRCA1 GTACCTTGATTTCGTATTCTGAGAGGCTGCTGCTTAGCGGTAGCCCCTTGGTTTCCGTGGCAACGGAAAA
How can we read this whole file in to a dictionary?
#!/usr/bin/env python3
seq_read = open("sequence_data.txt","r")
genes = {}
for line in seq_read:
line = line.rstrip()
id,seq = line.split() #split on whitespace
genes[id] = seq
seq_read.close()
print(genes)Output:
{'TP53': 'GATGGGATTGGGGTTTTCCCCTCCCATGTGCTCAAGACTGGCGCTAAAAGTTTTGAGCTTCTCAAAAGTC', 'BRCA1': 'GTACCTTGATTTCGTATTCTGAGAGGCTGCTGCTTAGCGGTAGCCCCTTGGTTTCCGTGGCAACGGAAAA'}
\pagebreak
Regular Expressions is a language for pattern matching. Many different computer languages incorporate regular expressions as well as some unix commands like grep and sed. So far we have seen a few functions for finding exact matches in strings, but this is not always sufficient.
Functions that utilize regular expressions allow for non-exact pattern matching.
These specialized functions are not included in the core of Python. We need to import them by typing
import re
at the top of your script
#!/usr/bin/env python3
import reFirst we will go over a few examples then go into the mechanics in more detail.
Let's start simple and find an exact match for the EcoRI restriction site in a string.
>>> dna = 'ACAAAATACGTTTTGTAAATGTTGTGCTGTTAACACTGCAAATAAACTTGGTAGCAAACACTTCCAAAAGGAATTCACCGGTTTCCAAAGACAGTCTTCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGGACCGGTTTCCAAAGACAGTCTTCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGG'
>>> if re.search(r"GAATTC",dna):
... print("Found an EcoRI site!")
...
Found an EcoRI site!
>>>Since we can search for control characters like a tab (\t) it is good to get in the habit of using the raw string function
rwhen defining patterns.
Here we used the
search()function with two arguments, 1) our pattern and 2) the string we want to search.
Let's find out what is returned by the search() function.
>>> dna = 'ACAAAATACGTTTTGTAAATGTTGTGCTGTTAACACTGCAAATAAACTTGGTAGCAAACACTTCCAAAAGGAATTCACCGGTTTCCAAAGACAGTCTTCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGGACCGGTTTCCAAAGACAGTCTTCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGG'
>>> found=re.search(r"GAATTC",dna)
>>> print(found)
<_sre.SRE_Match object; span=(70, 76), match='GAATTC'>Information about the first match is returned
How about a non-exact match. Let's search for a methylation site that has to match the following criteria:
- G or A
- followed by C
- followed by one of anything or nothing
- followed by a G
This could match any of these:
GCAG
GCTG
GCGG
GCCG
GCG
ACAG
ACTG
ACGG
ACCG
ACG
We could test for each of these, or use regular expressions. This is exactly what regular expressions can do for us.
>>> dna = 'ACAAAATACGTTTTGTAAATGTTGTGCTGTTAACACTGCAAATAAACTTGGTAGCAAACACTTCCAAAAGGAATTCACCGGTTTCCAAAGACAGTCTTCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGGACCGGTTTCCAAAGACAGTCTTCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGG'
>>> found=re.search(r"[GA]C.?G",dna)
>>> print(found)
<_sre.SRE_Match object; span=(7, 10), match='ACG'>Here you can see in the returned information that ACG starts at string postion 7 (nt 8). The first position following the end of the match is at string postion 10 (nt 11).
What about other potential matches in our DNA string? We can use findall() function to find all matches.
>>> dna = 'ACAAAATACGTTTTGTAAATGTTGTGCTGTTAACACTGCAAATAAACTTGGTAGCAAACACTTCCAAAAGGAATTCACCGGTTTCCAAAGACAGTCTTCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGGACCGGTTTCCAAAGACAGTCTTCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGG'
>>> found=re.findall(r"[GA]C.?G",dna)
>>> print(found)
['ACG', 'GCTG', 'ACTG', 'ACCG', 'ACAG', 'ACCG', 'ACAG']
findall()returns a list of all the pieces of the string that match the regex.
A quick count of all the matching sites can be done by counting the length of the returned list.
>>> len (re.findall(r"[GA]C.?G",dna))
7There are 7 methylation sites. Here we have another example of nesting. We call the
findall()function, searching for all the matches of a methylation site. This function returns a list, the list is past to thelen()function, which in turn returns the number of elements in the list.
Let's talk a bit more about all the new characters we see in the pattern.
The pattern in made up of atoms. Each atom represents ONE character.
| Atom | Description |
|---|---|
| a-z, A-Z, 0-9 and some punctuation | These are ordinary characters that match themselves |
| "." | The dot, or period. This matches any single character except for the newline. |
A group of characters that are allowed to be matched one time. There are a few predefined classes, symbol that means a series of characters.
| Atom | Description |
|---|---|
[ ] |
A bracketed list of characters, like [GA]. This indicates a single character can match any character in the bracketed list. |
\d |
Digits. Also can be written [0-9] |
\D |
Not digits. Also can be written[^0-9] |
\w |
Word character. Also can be written [A-Za-z0-9_] Note underscore is part of this class |
\W |
Not a word character, or [^A-Za-z0-9_] |
\s |
White space character. Also can be written [ \r\t\n]. Note the space character after the first [ |
\S |
Not whitespace. Also [^ \r\\t\n] |
A pattern can be anchored to a region in the string:
| Atom | Description |
|---|---|
^ |
Matches the beginning of the string |
$ |
Matches the end of the string |
\b |
Matches a word boundary between \w and \W |
Examples:
g..t
matches "gaat", "goat", and "gotta get a goat" (twice)
g[gatc][gatc]t
matches "gaat", "gttt", "gatt", and "gotta get an agatt" (once)
\d\d\d-\d\d\d\d`
matches 376-8380, and 5128-8181 but not 055-98-2818.
^\d\d\d-\d\d\d\d
matches 376-8380 and 376-83801 but not 5128-8181.
^\d\d\d-\d\d\d\d$
only matches telephone numbers (without area code)
\bcat
matches "cat", "catsup" and "more catsup please" but not "scat".
\bcat\b
only text containing the word "cat".
Quantifiers quantify how many atoms are to be found. By default an atom matches only once. This behaviour can be modified following an atom with a quantifier.
| Quantifier | Description |
|---|---|
? |
atom matches zero or exactly once |
* |
atom matches zero or more times |
+ |
atom matches one or more times |
{3} |
atom matches exactly 3 times |
{2,4} |
atom matches between 2 and 4 times, inclusive |
{4,} |
atom matches at least 4 times |
Examples:
goa?t
matches "goat" and "got". Also any text that contains these words.
g.+t
matches "goat", "goot", and "grant", among others.
g.*t
matches "gt", "goat", "goot", and "grant", among others.
^\d{3}-\d{4}$
matches US telephone numbers (no extra text allowed).
Something to think about.
- What would be a pattern to recognize an email address?
- What would be a pattern to recognize the ID portion of a sequence record in a FASTA file?
Variables can be used to store patterns.
>>> pattern = r"[GA]C.?G"
>>> len (re.findall(pattern,dna))
7In this example we stored our methylation pattern in the variable named 'pattern' and used it as the first argument to
findall.
A pipe '|' can be used to indicated that either the pattern before or after the '|' can match. Enclose the two options in parenthesis.
big bad (wolf|sheep)
This pattern must match a string that contains "big" followed by a space followed by "bad" followed by a space followed by either "wolf" or "sheep" This would match, "big bad wolf" Or "big bad sheep"
Something to think about.
- What would a pattern to match 'ATG' followed by a C or a T look like?
Subpatterns, or parts of the pattern enclosed in parenthesis can be extracted and stored for later use.
Who's afraid of the big bad w(.+)f
This pattern has only one subpattern (.+)
You can combine parenthesis and quantifiers to quantify entire subpatterns.
Who's afraid of the big (bad )?wolf\?
This matches "Who's afraid of the big bad wolf?". As well as "Who's afraid of the big wolf?". The 'bad ' is optional, it can be present 0 or 1 times in our string. This also shows how to literally match special characters. Use a '' in to escape them.
Something to think about: How would you create a pattern to capture the ID in a sequence record of a FASTA file in a subpattern.
Example FASTA sequence record.
>ID Optional Descrption
SEQUENCE
SEQUENCE
SEQUENCE
This is helpful when you want to find a subpattern and then match the contents again. They can be used within the function call and used after the function call.
Subpatterns within the function call
Once a subpattern matches, you can refer to it within the same regular expression. The first subpattern becomes \1, the second \2, the third \3, and so on.
Who's afraid of the big bad w(.)\1f
This would match "Who's afraid of the big bad woof" "Who's afraid of the big bad weef" "Who's afraid of the big bad waaf" But Not, "Who's afraid of the big bad wolf" Or, "Who's afraid of the big bad wife"
In a similar vein,
\b(\w+)s love \1 food\b
This pattern will match "dogs love dog food" But not "dogs love monkey food". We were able to use the subpattern within the regular expression by using
\1If there were more subpatterns they would be\2,\3,\4, etc
Subpatterns can be retrieved after the search() function call, or outside the regular expression, by using the group() method. This is a method and it belongs to the object that is returned by the search() function.
The subpatterns are retrieved by a number. This will be the same number that could be used within the regular expression, i.e.,
\1within the subpattern can be used outside withsearch_found_obj.group(1)\2within the subpattern can be used outside withsearch_found_obj.group(2)\3within the subpattern can be used outside withsearch_found_obj.group(3)- and so on
Example:
>>> dna = 'ACAAAATACGTTTTGTAAATGTTGTGCTGTTAACACTGCAAATAAACTTGGTAGCAAACACTTCCAAAAGGAATTCACCGGTTTCCAAAGACAGTCTTCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGGATATTATCCGGTTTCCAAAGACAGTCTTCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGG'
>>> found=re.search( r"(.{50})TATTAT(.{25})" , dna )
>>> upstream = found.group(1))
>>> print(upstream)
TCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGGA
>>> downstream = found.group(2))
>> print(downstream)
CCGGTTTCCAAAGACAGTCTTCTAA
- This pattern will recognize a consensus transcription start site (TATTAT)
- And store the 50 base pairs upstream of the site
- And the 25 base pairs downstream of the site
If you want to find the upstream and downstream sequence of ALL 'TATTAT' sites, use the findall() function.
>>> dna="ACAAAATACGTTTTGTAAATGTTGTGCTGTTAACACTGCAAATAAACTTGGTAGCAAACACTTCCAAAAGGAATTCACCGGTTTCCAAAGACAGTCTTCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGGATATTATCCGGTTTCCAAAGACAGTCTTCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGGACAAAATACGTTTTGTAAATGTTGTGCTGTTAACACTGCAAATAAACTTGGTAGCAAACACTTCCAAAAGGAATTCACCGGTTTCCAAAGACAGTCTTCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGGATATTATCCGGTTTCCAAAGACAGTCTTCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGG"
>>> found = re.findall( r"(.{50})TATTAT(.{25})" , dna )
>>> print(found)
[('TCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGGA', 'CCGGTTTCCAAAGACAGTCTTCTAA'), ('TCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGGA', 'CCGGTTTCCAAAGACAGTCTTCTAA')]The subpatterns are stored in tuples within a list. More about this type of data structure later.
Another option for retrieving the upstream and downstream subpatterns is to put the findall() in a for loop
>>> dna="ACAAAATACGTTTTGTAAATGTTGTGCTGTTAACACTGCAAATAAACTTGGTAGCAAACACTTCCAAAAGGAATTCACCGGTTTCCAAAGACAGTCTTCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGGATATTATCCGGTTTCCAAAGACAGTCTTCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGGACAAAATACGTTTTGTAAATGTTGTGCTGTTAACACTGCAAATAAACTTGGTAGCAAACACTTCCAAAAGGAATTCACCGGTTTCCAAAGACAGTCTTCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGGATATTATCCGGTTTCCAAAGACAGTCTTCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGG"
>>> for (upstream, downstream) in re.findall( r"(.{50})TATTAT(.{25})" , dna ):
... print("upstream:" , upstream)
... print("downstream:" , downstream)
...
upstream: TCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGGA
downstream: CCGGTTTCCAAAGACAGTCTTCTAA
upstream: TCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGGA
downstream: CCGGTTTCCAAAGACAGTCTTCTAA
- This code executes the
findall()function once- The subpatterns are returned
- The subpatterns are stored in the variables upstream and downstream
- The for block of code is executed
- The
findall()searches again- A match is found
- New subpatterns are returned and stored in the variables upstream and downstream
- The for block of code gets executed again
- The
findall()searches again, but no match is found- The for loop ends
Another way to get this done is with an iterator, use the finditer() function in a for loop. This allows you to not store all the matches in memory. finditer() also allows you to retrieve the postion of the match.
>>> dna="ACAAAATACGTTTTGTAAATGTTGTGCTGTTAACACTGCAAATAAACTTGGTAGCAAACACTTCCAAAAGGAATTCACCGGTTTCCAAAGACAGTCTTCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGGATATTATCCGGTTTCCAAAGACAGTCTTCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGGACAAAATACGTTTTGTAAATGTTGTGCTGTTAACACTGCAAATAAACTTGGTAGCAAACACTTCCAAAAGGAATTCACCGGTTTCCAAAGACAGTCTTCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGGATATTATCCGGTTTCCAAAGACAGTCTTCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGG"
>>> for match in re.finditer(r"(.{50})TATTAT(.{25})" , dna):
... print("upstream:" , match.group(1))
... print("downstream:" , match.group(2))
...
upstream: TCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGGA
downstream: CCGGTTTCCAAAGACAGTCTTCTAA
upstream: TCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGGA
downstream: CCGGTTTCCAAAGACAGTCTTCTAA
- This code executes
finditer()function once.- The match object is returned. A match object will have all the information about the match
- In the for block we call the
group()method on the first match object returned- We print out the first and second subpattern using the
group()method- The
finditer()function is executed a second time and a match is found- The second match object is returned
- The second subpatterns are retrieved from the match object using the
group()method- The
finditer()function is executed again, but no matches found, so the loop ends
The match object contains information about the match that can be retrieved with match methods like start() and end()
#!/usr/bin/env python3
import re
dna="ACAAAATACGTTTTGTAAATGTTGTGCTGTTAACACTGCAAATAAACTTGGTAGCAAACACTTCCAAAAGGAATTCACCGGTTTCCAAAGACAGTCTTCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGGATATTATCCGGTTTCCAAAGACAGTCTTCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGGACAAAATACGTTTTGTAAATGTTGTGCTGTTAACACTGCAAATAAACTTGGTAGCAAACACTTCCAAAAGGAATTCACCGGTTTCCAAAGACAGTCTTCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGGATATTATCCGGTTTCCAAAGACAGTCTTCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGG"
for found in re.finditer(r"(.{50})TATTAT(.{25})" , dna):
whole = found.group(0)
up = found.group(1)
down = found.group(2)
up_start = found.start(1) + 1 # need to convert from 0 to 1 notation
up_end = found.end(1) + 1
dn_start = found.start(2) + 1
dn_end = found.end(2) + 1
output = [ whole , up , str(up_start), str(up_end) , down , str(dn_start) , str(dn_end) ]
print( "\t".join(output) )we can use these match object methods
group(),start(),end()to get the string, start position, and end position of each subpattern.
$ python3 re.finditer.pos.py
TCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGGATATTATCCGGTTTCCAAAGACAGTCTTCTAA TCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGGA 98 148 CCGGTTTCCAAAGACAGTCTTCTAA 154 179
TCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGGATATTATCCGGTTTCCAAAGACAGTCTTCTAA TCTAATTCCTCATTAGTAATAAGTAAAATGTTTATTGTTGTAGCTCTGGA 320 370 CCGGTTTCCAAAGACAGTCTTCTAA 376 401
FYI: match() function is another regular expression function that looks for patterns. It is similar to search but it only looks at the beginning of the string for the pattern while search() looks in the entire string. Usually finditer() , search() and findall() will be more useful.
By default, regular expressions are "greedy". They try to match as much as they can. Use the quantifier '?' to make the match not greedy. The not greedy match is called 'lazy'
>>> str = 'The fox ate my box of doughnuts'
>>> found = re.search(r"(f.+x)",str)
>>> print(found.group(1))
fox ate my boxThe pattern f.+x does not match what you might expect, it matches past 'fox' all the way out to 'fox ate my box'.
The '.+' id greedy As many characters as possible are found that are between the 'f' and the 'x'.
Let's make this match lazy by using '?'
>>> found = re.search(r"(f.+?x)",str)
>>> print(found.group(1))
foxThe match is now lazy and will only match 'fox'
Extracting codons from a string of DNA can be accomplished by using a subpattern in a findall() function. Remember the findall() function will return a list of the matches.
>>> dna = 'GTTGCCTGAAATGGCGGAACCTTGAA'
>>> codons = re.findall(r"(.{3})",dna)
>>> print(codons)
['GTT', 'GCC', 'TGA', 'AAT', 'GGC', 'GGA', 'ACC', 'TTG']Or you can use a for loop to do something to each match.
>>> for codon in re.findall(r"(.{3})",dna):
... print(codon)
...
GTT
GCC
TGA
AAT
GGC
GGA
ACC
TTG
>>>
finditer()would also work in this for loop.
Each codon can be accessed by using thegroup()method.
The search(), match(), findall(), and finditer() can be used in conditional tests. If a match is not found an empty list or 'None' is returned. These both are False.
>>> found=re.search( r"(.{50})TATTATZ(.{25})" , dna )
>>> if found:
... print("found it")
... else:
... print("not found")
...
not found
>>> print(found)
NoneNone is False so the else block is executed and "not found" is printed
Nest it!
>>>
>>> if re.search( r"(.{50})TATTATZ(.{25})" , dna ):
... print("found it")
... else:
... print("not found")
...
not found
>>> print(found)
NoneUsing Regular expressions in substitutions
Earlier we went over how to find an exact pattern and replace it using the replace() method. To find a pattern, or inexact match, and make a replacement the regular expression sub() function is used. This function takes the pattern, the replacement, the string to be searched, the number of times to do the replacement, and flags.
>>> str = "Who's afraid of the big bad wolf?"
>>> re.sub(r'w.+f' , 'goat', str)
"Who's afraid of the big bad goat?"
>>> print(str)
Who's afraid of the big bad wolf?The
sub()function returns "Who's afraid of the big bad goat?"
The value of variable str has not been altered
The new string can be stored in a new variable for later use.
Let's save the returned new string in a variable
>>> str = "He had a wife."
>>> new_str = re.sub(r'w.+f' , 'goat', str)
>>> print(new_str)
He had a goate.
>>> print(str)
He had a wife.The characters between 'w' and 'f' have been replaced with 'goat'.
The new string is saved in new_str
Sometimes you want to find a pattern and use it in the replacement.
>>> str = "Who's afraid of the big bad wolf?"
>>> new_str = re.sub(r"(\w+) (\w+) wolf" , r"\2 \1 wolf" , str)
>>> print(new_str)
Who's afraid of the bad big wolf?We found two words before 'wolf' and swapped the order. \2 refers to the second subpattern \1 refers to the first subpattern
Something to think about.
How would you use regular expressions to find all occurrences of 'ATG' and replace with '-M-' in this sequence 'GCAGAGGTGATGGACTCCGTAATGGCCAAATGACACGT'?
| Modifier | Description |
|---|---|
re.I re.IGNORECASE |
Performs case-insensitive matching. |
re.M re.MULTILINE |
Makes $ match the end of a line (not just the end of the string) and makes ^ match the start of any line (not just the start of the string). |
re.S re.DOTALL |
Makes a period (dot) match any character, including a newline. |
re.U |
Interprets letters according to the Unicode character set. This flag affects the behavior of \w, \W, \b, \B. |
re.X VERBOSE |
This flag allows you to write regular expressions that look nicer and are more readable by allowing you to visually separate logical sections of the pattern and add comments. Whitespace within the pattern is ignored, except when in a character class or when preceded by an unescaped backslash. When a line contains a # that is not in a character class and is not preceded by an unescaped backslash, all characters from the leftmost such # through the end of the line are ignored. |
>>> dna = "atgcgtaatggc"
>>> re.search(r"ATG",dna)
>>>
>>> re.search(r"ATG",dna , re.I)
<_sre.SRE_Match object; span=(0, 3), match='atg'>
>>>We can make our search case insensitive by using the
re.Iorre.IGNORECASEflag.
You can use more than one flag by concatenating them with |. re.search(r"ATG",dna , re.I|re.M)
Functions consist of lines of code that do something useful and that you want to run more than once. You also give that function a name so you can refer to it in your code. This avoids copying and pasting the code to many places in your script and makes your code easier to read.
Let's see some examples.
Python has built-in functions
>>> print('Hello world!')
Hello world!
>>> len('AGGCT')
5You can also define your own functions with def Let's write a function that calculates the GC content. Let's define this as the fraction of nucleotides in a DNA sequence that are G or C. It can vary from 0 to 1.
First we can look at the code that makes the calculation, then we can convert those lines of code into a function.
Code to find GC content:
dna = 'GTACCTTGATTTCGTATTCTGAGAGGCTGCTGCT'
c_count = dna.count('C') # count is a string method
g_count = dna.count('G')
dna_len = len(dna) # len is a function
gc_content = (c_count + g_count) / dna_len # fraction from 0 to 1
print(gc_content)We use def do define our own function. It is followed by the name of the function (gc_content) and parameters it will take in parentheses. A colon is the last character on the def line. The parameter variables will be available for your code inside the function to use.
def gc_content(dna): # give our function a name and parameter 'dna'
c_count = dna.count('C')
g_count = dna.count('G')
dna_len = len(dna)
gc_content = (c_count + g_count) / dna_len
return gc_content # return the value to the code that called this functionHere is a custom function that you can use like a built in Python function
This is just like any other python function. You write the name of the function with any variables you want to pass to the function in parentheses. In the example below the contents of dna_string get passed into gc_content(). Inside the function this data is passed to the variable dna.
dna_string = "GTACCTTGATTTCGTATTCTGAGAGGCTGCT"
print(gc_content(dna_string))This code will print 0.45161290322580644 to the screen. You can save this value in a variable to use later in your code like this
dna_gc = gc_content('GTACCTTGATTTCGTATTCTGAGAGGCTGCT')As you can see we can write a nice clear line of python to call this function and because the function has a name that describes what it does it's easy to understand how the code works. Don't define your functions like this def my_function(a):!
How could you convert the GC fraction to % GC. Use format()
dna_string = "GTACCTTGATTTCGTATTCTGAGAGGCTGCT"
dna_gc = gc_content(dna_string)
pc_gc = '{:.2%}'.format(dna_gc)
print('This sequence is' , pc_gc , 'GC')Here's the output
This sequence is 45.16% GC- You define a function with
def. You need to define a function before you can call it. - The function must have a name. This name should clearly describe what the function does. Here is our example
gc_content - You can pass variables to functions but you don't have to. In the definition line, you place variables your function needs inside parentheses like this
(dna). This variable only exists inside the function. - The first line of the function must end with a
:so the complete function definition line looks like thisdef gc_content(dna): - The next lines of code, the function body, needs to be indented. This code comprises what the function does.
- You can return a value as the last line of the function, but this is not required. This line
return gc_contentat the end of our function definition passes the value of gc_content back to the code that called the function in your main script.
You can name your argument variables anything you want, but they should describe the data they contain. The name needs to be consistent within your function. You could change dna to seqeunce like this
def gc_content(sequence): # give our function a name and parameter 'sequence'
c_count = sequence.count('C')
g_count = sequence.count('G')
dna_len = len(sequence)
gc_content = (c_count + g_count) / dna_len
return gc_content # return the value of gc_content to the code that called this functionArguments can be named and these names can be used when the function is called. This name is called a 'keyword'
>>> dna_string = "GTACCTTGATTTCGTATTCTGAGAGGCTGCT"
>>> print(gc_content(dna_string))
0.45161290322580644
>>> print(gc_content(dna=dna_string)
0.45161290322580644The keyword must be the same as the defined function argument. If a function has multiple arguments, using the keyword allows for calling the function with the arguments in any order.
As defined above, our function is expecting an argument (dna) in the definition. You get an error if you call the function without any parameters.
>>> gc_content()
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
TypeError: gc_content() missing 1 required positional argument: 'dna'You can define default values for arguments when you define your function.
def gc_content(dna='A'): # give our function a name and parameter 'dna'
c_count = dna.count('C')
g_count = dna.count('G')
dna_len = len(dna)
gc_content = (c_count + g_count) / dna_len
return gc_content # return the value to the code that called this functionIf you call the function with no arguments, the default will be used. In this case a default is pretty useless, and the function will return '0' if called without providing a DNA sequence.
Lambda expressions can be used to define simple (one-line) anonymous functions. There are some uses for lambda which we won't go into here. We are showing it to you because sometimes you will come across it.
Here is a one line custom function, like the functions we have already talked about:
def get_first_codon(dna):
return dna[0:3]
print(get_first_codon('ATGTTT'))This will print
ATG
Here is the same function written as a lambda
get_first_codon = lambda dna : dna[0:3]
print(get_first_codon('ATGTTT'))This also prints
ATG. lambdas can only contain one line and there is noreturnstatement.
List comprehensions can often be used instead of lambdas and may be easier to read. You can read more about lambda, particularly in relation to map which will perform an operation on a list, but generally a for loop is easier to read.
Almost all python variables are global. This means they are available everywhere in your code. The most important exception is variables thare are defined in functions which only exist inside their function. This is called 'local'. Remember that python blocks are defined as code at the same level of indentation.
#!/usr/bin/env python3
print('Before if block')
x = 100
print('x=',x)
if True: # this if condition will always be True
# we want to make sure the block gets executed
# so we can show you what happens
print('Inside if block')
y = 10
x = 30
print("x=", x)
print("y=", y)
print('After if block')
print("x=", x)
print("y=", y)
Let's Run it:
$ python3 scripts/scope.py
Before if block
x= 100
Inside if block
x= 30
y= 10
After if block
x= 30
y= 10
Inside a function, global variables are visible, but it's better to pass variables to a function as arguments
def show_n():
print(n)
n = 5
show_n()The output is this 5 as you would expect, but this is better programming practice. Why? We'll see a little later.
def show_n(n):
print(n)
n = 5
show_n(n)Variables inside functions are local and therefore can only been accessed from within the function block. This applies to arguments as well as variables defined inside a function.
#!/usr/bin/end python3
def set_local_x_to_five(x):
print('Inside def')
x = 5 # local to set_local_x_to_five()
y=5
print("x =",x)
print("y = ",y)
print('After def')
x = 100 # global x
y = 100 # global
print('x=',x)
print('y=',y)
set_local_x_to_five(500)
print('After function call')
print('x=',x)
print('y=',y)Here we have added a function
set_local_x_to_fivewith an argument named 'x'. This variable exists only within the function where is replaces any variable with the same name outside thedef. Inside thedefwe also initialize a variableythat also replaces any globalywithin thedef
Let's run it:
$ python3 scope_w_function.py
After def
x= 100
y= 100
Inside def
x = 5
y = 5
After function call
x= 100
y= 100
There is a global variable,
x= 100, but when the function is called, it makes a new local variable, also calledxwith value = 5. This variable disappears after the function finishes and we go back to using the global variablex= 100. Same fory
You can make a local variable global with the statement global. Now a variable you use in a function is the same variable as in the rest of the code. It is best not to define any variables as global until you know you need to because you might modify the contents of a variable without meaning to.
Here is an example use of global.
#!/usr/bin/env python3
def set_global_variable():
global greeting # make greeting global
greeting = "I say hello"
greeting = 'Good morning'
print('Before function call')
print('greeting =',greeting)
#make call to function
set_global_variable()
print('After function call')
print('greeting =',greeting)Let's look at the output
$ python3 scripts/scope_global.py
Before function call
greeting = Good morning
After function call
greeting = I say hello
Note that the function has changed the value of the global variable. You might not want to do this.
Python comes with some core functions and methods. There are many useful modules that you will want to use. import is the statement for telling your script you want to use code in a module. As we've already seen with regular expresions, you can bring in code that handles regular expressions with import re
How do you find out information about a module? Python has help pages built into the command line, like man we met earlier in the unix lecture. Online information may be more up to date. Search at https://docs.python.org/3.6/. But if you don't have internet access, you can always use pydoc.
To find out about the re module, type pydoc re on the command line. The last line in the output tells you where the python module is actually installed.
% pydoc re
Help on module re:
NAME
re - Support for regular expressions (RE).
MODULE REFERENCE
https://docs.python.org/3.6/library/re
The following documentation is automatically generated from the Python
source files. It may be incomplete, incorrect or include features that
are considered implementation detail and may vary between Python
implementations. When in doubt, consult the module reference at the
location listed above.
DESCRIPTION
This module provides regular expression matching operations similar to
those found in Perl. It supports both 8-bit and Unicode strings; both
the pattern and the strings being processed can contain null bytes and
characters outside the US ASCII range.
Regular expressions can contain both special and ordinary characters.
Most ordinary characters, like "A", "a", or "0", are the simplest
regular expressions; they simply match themselves. You can
concatenate ordinary characters, so last matches the string 'last'.
...
FILE
/anaconda3/lib/python3.6/glob.py
Here are some of the most common and useful modules, along with their methods and objects. It's a lightning tour.
os.path has common utilities for working file paths (filenames and directories). A path is either a relative or absolute list of directories (often ending with a filename) that tells you where to find a file or directory.
| function | description |
|---|---|
| os.path.basename(path) | what's the last element of the path? Note /home/tmp/ returns '', rather than tmp |
| os.path.dirname(path) | what's the directory the file is in? |
| os.path.exists(path) | does the path exist? |
| os.path.getsize(path) | returns path (file) size in bytes or error |
| os.path.isfile(path) | does the path point to a file? |
| os.path.isdir(path) | does the path point to a directory? |
| os.path.splitext(path) | splits before and after the file extension (e.g. '.txt') |
Run a system command from python. This is like making a python script run something from the command line. Replaced by subprocess
import os
os.system("ls -l")updated module for running command lines from python scripts
import subprocess
subprocess.run(["ls","-l"]) # same as running ls -l on the command linemore complex than os.system(). You need to specify where input and output go. Let's look at this in some more detail.
Let's say we want to find all the files that have user amanda (or in the filename)
ls -l | grep amanda
becomes this 'shortcut' which will capture the output of the two unix commands in the variable output
import subprocess
output = subprocess.check_output('ls -l | grep amanda', shell = True)This is better than alternatives with subprocess.run(). This is equivalent to the unix backtick quoted string.
output contains a bytes object (more or less a string of ASCII character encodings)
b'-rw-r--r-- 1 amanda staff 161952 Oct 2 18:03 test.subreads.fa\n-rw-r--r-- 1 amanda staff 126 Oct 2 13:23 test.txt\n'You can covert by decoding the bytes object into a string
>>>output.decode('utf-8')
'-rw-r--r-- 1 amanda staff 161952 Oct 2 18:03 test.subreads.fa\n-rw-r--r-- 1 amanda staff 126 Oct 2 13:23 test.txt\n'Let's assume that ls -l generates some output something like this
total 112
-rw-r--r-- 1 amanda staff 69 Jun 14 17:41 data.cfg
-rw-r--r-- 1 amanda staff 161952 Oct 2 18:03 test.subreads.fa
-rw-r--r-- 1 amanda staff 126 Oct 2 13:23 test.txt
How do we run ls -l in Python and capture the output (stdout)?
import subprocess
rtn = subprocess.run(['ls','-l'], stdout=subprocess.PIPE ) # specify you want to capture STDOUT
bytes = rtn.stdout
stdout = bytes.decode('utf-8')
# something like
lines = stdout.splitlines()lines now contains elements from every line of the ls -l output, including the header line, which is not a file
>>> lines[0]
'total 112'
>>> lines[1]
'-rw-r--r-- 1 amanda staff 69 Jun 14 17:41 data.cfg'To run a command and check the exit status (really to check the exit status was ok or zero), use
oops = subprocess.check_call(['ls', '-l'])
# or, simpler...
oops = subprocess.check_call('ls -l', shell=True)You can't write ls -l > listing.txt to redirect stdout in the subprocess method, so use this instead
tmp_file = 'listing.txt'
with open(tmp_file,'w') as ofh:
oops = subprocess.check_call(['ls', '-l'], stdout=ofh )A couple of useful variables for beginners. Many more advanced system parameters and settings that we are not covering here.
| function | description |
|---|---|
| sys.argv | list of command line parameters |
| sys.path | where Python should look for modules |
See notes on regular expressions
Better lists etc.
from collections import deque
copy.copy()
and
copy.deepcopy()
| function | description |
|---|---|
| math.exp() | e**x |
| math.log2() | log base 2 |
| math.log10() | log base 10 |
| math.sqrt() | square root |
| math.sin() | sine |
| math.pi(), math.e() | constants |
| etc |
see also numpy
Random numbers generated by computers are not truly random, so python calls these pseudo-random.
| example | description |
|---|---|
| random.seed(1) | set starting seed for random sequence to 1 to enable reproducibility |
| random.randrange(9) | integer between 0 and 8 |
| random.randint(1,5) | integer between 1 and 5 |
| random.random() | float between 0 and 1 |
| random.uniform(1,2) | float between 1 and 2 |
To get a random index from an element of list use i=random.randrange(len(list))
Typical statistical quantities
| example | description |
|---|---|
| statistics.mean([1,2,3,4,5]) | mean or average |
| statistics.median([ 2,3,4,5]) | median = 3.5 |
| statistics.stdev([1,2,3,4,5]) | standard deviation of sample (square root of sample variance) |
| statistics.pstdev([1,2,3,4,5])q | estimate of population standard deviation |
Does unix-like wildcard file path expansion.
>>> import glob
>>> glob.glob('pdfs/*.pdf')
['pdfs/python1.pdf', 'pdfs/python2.pdf', 'pdfs/python3.pdf', 'pdfs/python4.pdf', 'pdfs/python6.pdf', 'pdfs/python8.pdf', 'pdfs/unix1.pdf', 'pdfs/unix2.pdf']
>>> fasta_files = glob.glob('sequences/*.fa')
>>> ```
#### argparse
Great (if quite complicated) tool for parsing command line arguments and automatically generating help messages for scripts (very handy!). Here's a simple script that explains a little of what it does.
```python
#!/usr/bin/env python3
import argparse
parser = argparse.ArgumentParser(description="A test program that reads in some number of lines from an input file. The output can be screen or an output file")
# we want the first argument to be the filename
parser.add_argument("file", help="path to input fasta filename")
# second argument will be line number
# default type is string, need to specify if expecting an int
parser.add_argument("lines", type=int, help ="how many lines to print")
# optional outfile argument specified with -o or --out
parser.add_argument("-o","--outfile", help = "optional: supply output filename, otherwise write to screen", dest = 'out')
args = parser.parse_args()
# arguments appear in args
filename = args.file
lines = args.line
if args.out:
print("writing output to", args.out)time, HTML, XML, email, CGI, sockets, audio, GUIs with Tk, debugging, testing, unix utils
Also, non-core: BioPython for bioinformatics, Numpy for mathematics, statistics
Classes allow us to create our own types of objects. They work just like the other objects we've met, like strings and lists and so on. In fact everything in python. Classes inherit functionality from other classes. This is an easy way to write code that quickly gains functions by re-using existing code. Classes are essentially packages of functions. You don't need to use these constructs, but sometimes they help in larger projects where there's more time to plan.
You can create objects that belong to a class. They get functions and attributes. Here's an example list.items
- create class
- make module with methods and attributes
Sometimes a simple list or dictionary just doesn't do what you want. Sometimes you need to organize data in a more complex way. You can nest any data type inside any other type. This lets you build multidimensional data tables easily.
List of lists, often called a matrix are important for organizing and accessing data
Here's a way to make a 3 x 3 table of values.
>>> M = [[1,2,3], [4,5,6],[7,8,9]]
>>> M[1] # second row (starts with index 0)
[4,5,6]
>>>M[1][2] # second row, third element
6Here's a way to store sequence alignment data:
Four sequences aligned:
AT-TG
AATAG
T-TTG
AA-TA
The alignment in a list of lists.
aln = [['A', 'T', '-', 'T', 'G'],
['A', 'A', 'T', 'A', 'G'],
['T', '-', 'T', 'T', 'G'],
['A', 'A', '-', 'T', 'A']]Get an the full length of one sequence:
>>> seq = aln[2]
>>> seq
['T', '-', 'T', 'T', 'G']Use the outer most index to access each sequence
Retrieve the nucleotide at a particular position in a sequence.
>>> nt = aln[2][3]
>>> nt
'T'Use the outer most index to access the sequence of interest and the inner most index to access the position
Get every nucleotide in a single column:
>>> col = [seq[3] for seq in aln]
>>> col
['T', 'A', 'T', 'T']Retrieve each sequence from the aln list then the 3rd column for each sequence.
You can nest dictionaries in lists as well:
>>> records = [
... {'name' : 'actgctagt', 'accession' : 'ABC123', 'genetic_code' : 1},
... {'name' : 'ttaggttta', 'accession' : 'XYZ456', 'genetic_code' : 1},
... {'name' : 'cgcgatcgt', 'accession' : 'HIJ789', 'genetic_code' : 5}
... ]
>>> records[0]['name']
'actgctagt'
>>> records[0]['accession']
'ABC123'
>>> records[0]['genetic_code']
1Here you can retrieve the accession of one record at a time by using a combination of the outer index and the key 'accession'
And, if you haven't guessed, you can nest lists in dictionaries
Here is a dictionary of kmers. The key is the kmer and its values is a list of postions
>>> kmers = {'ggaa': [4, 10], 'aatt': [0, 6, 12], 'gaat': [5, 11], 'tgga':
... [3, 9], 'attg': [1, 7, 13], 'ttgg': [2, 8]}
>>> kmers
{'tgga': [3, 9], 'ttgg': [2, 8], 'aatt': [0, 6, 12], 'attg': [1, 7, 13], 'ggaa': [4, 10], 'gaat': [5, 11]}
>>>
>>> kmers['ggaa']
[4, 10]
>>> len(kmers['ggaa'])
2Here we can get a list of the positions of a kmer by using the kmer as the key. We can also do things to the returned list, like determining its length. The length will be the total count of this kmers.
You can also use the get() method to retrieve records.
>>> kmers['ggaa']
[4, 10]
>>> kmers.get('ggaa')
[4, 10]These two statements returns the same results, but if the key does not exist you will get nothing and not an error.
Dictionaries of dictionaries is my favorite!! You can do so many useful things with this data structure. Here we are storing a gene name and some different types of information about that gene, such as its, sequence, length, description, nucleotide composition and length.
>>> genes = {
... 'gene1' : {
... 'seq' : "TATGCC",
... 'desc' : 'something',
... 'len' : 6,
... 'nt_comp' : {
... 'A' : 1,
... 'T' : 2,
... 'G' : 1,
... 'C' : 2,
... }
... },
...
... 'gene2' : {
... 'seq' : "CAAATG",
... 'desc' : 'something',
... 'len' : 6,
... 'nt_comp' : {
... 'A' : 3,
... 'T' : 1,
... 'G' : 1,
... 'C' : 1,
... }
... }
... }
>>> genes
{'gene1': {'nt_comp': {'C': 2, 'G': 1, 'A': 1, 'T': 2}, 'desc': 'something', 'len': 6, 'seq': 'TATGCC'}, 'gene2': {'nt_comp': {'C': 1, 'G': 1, 'A': 3, 'T': 1}, 'desc': 'something', 'len': 6, 'seq': 'CAAATG'}}
>>> genes['gene2']['nt_comp']
{'C': 1, 'G': 1, 'A': 3, 'T': 1}Here we store a gene name as the outermost key, with a second level of keys for qualities of the gene, like sequence, length, nucleotide composition. We can retrieve a quality by using the gene name and quality in conjunction.
There are also specific data table and frame handling libraries like Pandas. Here is a intro to data structures in Panda.
There are a few different types of errors when coding. Syntax errors, logic errors, and exceptions. You have probably encountered all three. Sytax and logic errors are issues you need to deal with while coding. An exception is a special type of error that can be informative and used to write code to respond to this type of error. This is especially relavent when dealing with user input. What if they don't give you any, or it is the wrong kind of input. We want our code to be able to detect these types of errors and respond accordingly.
#!/usr/bin/env python3
import sys
file = sys.argv[1]
print("User provided file:" , file)This code takes user provided input and prints it
Run it.
$ python scripts/exceptions.py test.txt
User provided file: test.txt
What happens if the user does not provide any input and we try to print it?
$ python scripts/exceptions.py
Traceback (most recent call last):
File "scripts/exceptions.py", line 4, in <module>
file = sys.argv[1]
IndexError: list index out of range We get an IndexError exception
We have already seen quite a few exceptions, here are some:
- ValueError: math domain error
- AttributeError: 'list' object has no attribute 'rstrip'
- SyntaxError: EOL while scanning string literal
- NameError: name 'GGTCTAC' is not defined
- SyntaxError: Missing parentheses in call to 'print'
- AttributeError: 'int' object has no attribute 'lower'
- IndexError: list assignment index out of range
- NameError: name 'HDAC' is not defined
We can use the exception to our advantage to help out our users. We can use a try/except condition to look for exceptions and to do something if we do not have an exception and do something different if we do have an exception.
#!/usr/bin/env python3
import sys
file = ''
try:
file = sys.argv[1]
print("User provided file:" , file)
except:
print("Please provide a file name")We need to "try" to get a user provided argument. If we are successful then we can print it out. If we try and fail, we execute the code in the except portion of our try/except and print that we need a file name.
Let's run it WITH iser input
$ python3 scripts/exceptions_try.py test.txt
User provided file: test.txtIt runs as expected
Let's run it WITHOUT user input
$ python scripts/exceptions_try.py
Please provide a file nameYeah, the user is informed that they need to provide a file name to the script
What if the user provides input but it is not a valid file or the path is incorrect? Or if you want to check to see if the user provided input as well as if it can open the input.
We can add multiple exception tests. Each except statement can specify what kind of exception it is waiting to recieve. If that kind of exception occures, that block of code will be executed.
import sys
file = ''
try:
file = sys.argv[1]
print("User provided file name:" , file)
FASTA = open(file, "r")
for line in FASTA:
line = line.rstrip()
print(line)
except IndexError:
print("Please provide a file name")
except IOError:
print("Can't find file:" , file)Here we test for an IndexError and a IOError. The IndexError occurs when we try to access a list element that does not exists. The IOError happens when we try to access a file that does not exist.
Let's run it with no input.
$ python scripts/exceptions_try_files.py test.txt
User provided file name: test.txt
Can't find file: test.txt
This informs the user that they did provide input but that the file listed can not be found.
Let's run it with no input
$ python scripts/exceptions_try_files.py
Please provide a file nameThis informs the user that they need to provide a file.
Lets summarize what we have covered and add on else and finally.
try:
# try block is executed until an exception is raised
except _ExceptionType_:
# if there is an exception of "ExceptionType" this block will be executed
# there can be more than one except block, just like an elif
except:
# if there are any exceptions that are not of _ExceptionType_ this except block will be executed
else:
# the else block is executed after the try block has been completed, which means there were no exceptions raised
finally:
# the finally block is executed if exceptions are or are not raised (no matter what happens)
Some exceptions can be thrown for multiple reasons, for example, ErrorIO will occur if the file does not exist as well as if you don't have permissions to read it. We can get more information by viewing the contents of our Exception Object. Yes, an exception is an object too! The system errors get stored in the exception object. To access the object use as and supply a variable name, like 'ex'
file = ''
try:
file = sys.argv[1]
print("User provided file name:" , file)
FASTA = open(file, "r")
for line in FASTA:
line = line.rstrip()
print(line)
except IndexError:
print("Please provide a file name")
except IOError as ex:
print("Can't find file:" , file , ': ' , ex.strerror ) Here we added
except IOError as exand now we can get the 'strerror' message from ex.
Run it.
$ python scripts/exceptions_try_files_as.py test.txt
User provided file name: test.txt
Can't find file: test.txt : No such file or directoryNow we know that this file name or path is not valid
We can call or raise exceptions too!! This is accomplished by using a raise statement.
- First, create a new Exception Object, i.e.,
ValueError() - Use the Exception Object in a Raise statment
raise ValueError('your message')
Let's raise an exception if the file name does not end in 'fa'
import sys
file = ''
try:
file = sys.argv[1]
print("User provided file name:" , file)
if not file.endswith('.fa'):
raise ValueError("Not a FASTA file")
FASTA = open(file, "r")
for line in FASTA:
print(line)
except IndexError:
print("Please provide a file name")
except IOError as ex:
print("Can't find file:" , file , ': ' , ex.strerror )Here we raise a known exception, 'ValueError', if the file does not end with (uses
endswith()method).
Let's run it.
$ python scripts/exceptions_try_files_raise.py test.txt
User provided file name: test.txt
Traceback (most recent call last):
File "scripts/exceptions_try_files_raise.py", line 10, in <module>
raise ValueError("Not a FASTA file")
ValueError: Not a FASTA fileOur exception get's raised, now lets do something with it.
import sys
file = ''
try:
file = sys.argv[1]
print("User provided file name:" , file)
if not file.endswith('.fa'):
raise ValueError("Not a FASTA file")
FASTA = open(file, "r")
for line in FASTA:
print(line)
except IndexError:
print("Please provide a file name")
except ValueError:
print("File needs to be a FASTA file and end with .fa")
except IOError as ex:
print("Can't find file:" , file , ': ' , ex.strerror )Here we created an except to catch any ValueError
Let's Run it.
$ python scripts/exceptions_try_files_raise_value.py test.txt
User provided file name: test.txt
File needs to be a FASTA file and end with .faWe get a great error message now.
But what if there is another ValueError, how can we tell if it is do to the FASTA file extension or not?
We can create our own custom exception. We will need to create a new class of exception. Below is the sytax to do this.
import sys
class NotFASTAError(Exception):
pass
file = ''
try:
file = sys.argv[1]
print("User provided file name:" , file)
if not file.endswith('.fa'):
raise NotFASTAError("Not a FASTA file")
FASTA = open(file, "r")
for line in FASTA:
print(line)
except IndexError:
print("Please provide a file name")
except NotFASTAError:
print("File needs to be a FASTA file and end with .fa")
except IOError as ex:
print("Can't find file:" , file , ': ' , ex.strerror )Here we created a new class of exception called 'NotFASTAError'. Then we raised this new exception.
Let's Run it.
$ python scripts/exceptions_try_files_raise_try.py test.txt
User provided file name: test.txt
File needs to be a FASTA file and end with .faOur new class of exception, NotFASTAError, works just like the built in exceptions.
- What you want to do:
- tools to do it