-
Notifications
You must be signed in to change notification settings - Fork 98
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
XBB.2.3 Sublineage with S:G184V, ORF1a:R2159W #1776
Comments
|
I was thinking to propose it too this morning with #1775 ! great you did find it and propose. |
|
Just took a closer look at the collection dates, and 18/34 sequences in this lineage have collection dates of March 1 or later, which is pretty remarkable and may indicate rapid growth. |
|
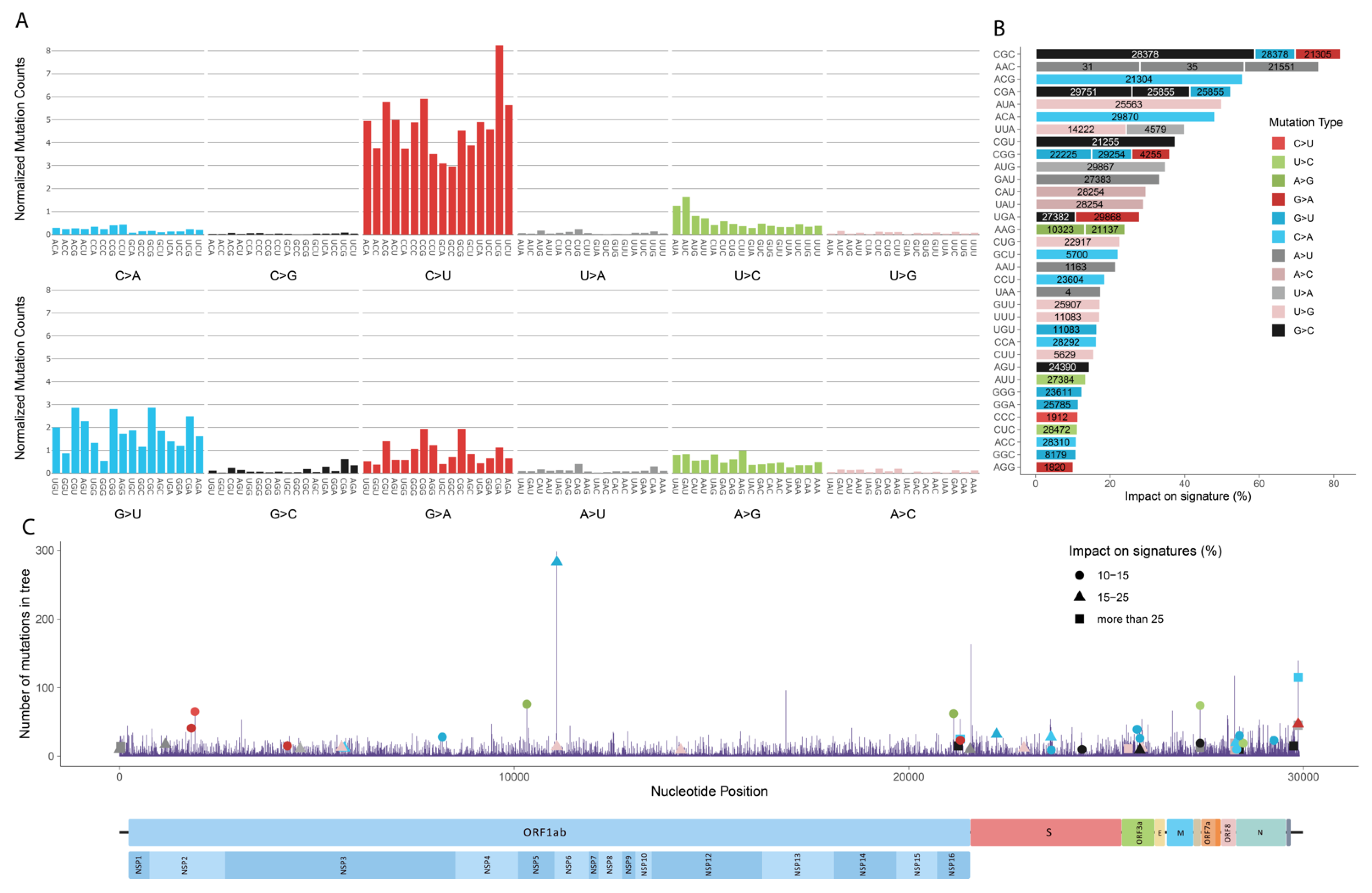
Interestingly, the result from this paper suggests that C>U mutations prefer G at the +1 position. |
|
Huh, that's interesting. I wasn't aware of that paper. Very different from the other one. @oobb45729, do you know of any other papers that measure the mutational context of C->T (and others)? I'd like to look at as many results as possible to see which of these two papers is more accurate as the results are very different. Bloom Lab showed that the rate of G->T mutations was roughly halved in Omicron compared to pre-Omicron lineages. I wonder if there is any difference in favored mutational contexts for different mutations. |
|
One paper counts from RNA sequencing datasets from bronchoalveolar lavage fluids obtained from patients diagnosed with COVID-19 while the other paper counts from a phylogenetic tree created from genome sequences from the GISAID. There's another paper Mutation rates and selection on synonymous mutations in SARS-CoV-2. |
|
56 as today big upload from Singapore Gujarat, one sequence from Guangdong. |
|
34 sequences uploaded today of this, 27 of them from Singapore. Also, could we get a milestone attached to this issue? Thanks. |
|
I did my own calculation based on the data from https://github.com/jbloomlab/SARS2-mut-fitness. |
|
Awesome, thanks for that analysis, @oobb45729! It's very interesting to contrast those findings with what we found in our preprint on molnupiravir sequences for C->T. • At +1: A favored, G very slightly favored, C disfavored, T highly disfavored Biggest contrast is G at -1, which has the largest positive effect of any preceding or following nucleotide in molnupiravir sequences but is disfavored in all other analyses. |

Description
Sub-lineage of: XBB.2.3
Earliest sequence: 2023-1-18, India, Gujarat — EPI_ISL_16743861
Most recent sequence: 2023-3-6, Singapore — EPI_ISL_17207252, EPI_ISL_17207254, EPI_ISL_17207255, EPI_ISL_17207257, EPI_ISL_17207258, EPI_ISL_17207261 (four local cases, two with no travel/local information)
Countries circulating: India (13), Singapore (13), USA (8)
Number of Sequences: 34
GISAID Query: C6740T, G22113T, T23018C
CovSpectrum Query: C6740T, G22113T, T23018C
Substitutions on top of XBB.2.3:
Spike: G184V
ORF1a: R2159W (NSP3_R1341W)
Nucleotide: C6740T, G22113T
USHER Tree

As usual, the long branches in the Usher tree below are due to artifactual reversions & should be disregarded.
https://nextstrain.org/fetch/raw.githubusercontent.com/ryhisner/jsons/main/XBB.2.3_G184V_ORF1aR2159W_subtreeAuspice1_genome_1ec30_5de800.json
Evidence
This lineage only very recently appeared and appears to be growing quickly. It seems to have originated in India and subsequently spread to Singapore and the US. Singapore uploads sequences faster than anyone, so I think this likely has spread elsewhere in Asia and will start to show up in sequences in the coming weeks.
Mutations from S:180 to S:186 have been ubiquitous in recent months, so it seems likely they confer a very modest increase in antibody evasion.
ORF1a:R2159W is more interesting. Overall, it has been a rare mutation throughout the pandemic. It was in an undesignated B.1 saltation lineage that rose to about 5% prevalence in Canada in late 2020/early 2021, and it was also in DN.1 (BQ.1.1.5 + S:K147N) and DN.1.1 (DN.1 + S:Y453F, ORF1a:V721I, ORF1a:N2752S). It involves a C->T mutation, which is easily the most common type. However, C->T mutation frequency is mostly driven by APOBEC, and the nucleotide context at C6740 (CA upstream and GG downstream) is unfavorable for APOBEC, which prefers A or T in the two closest upstream & downstream nucleotides.


Image below from "Evidence for host-dependent RNA editing in the transcriptome of SARS-CoV-2," by Di Giorgi, et al.
https://www.science.org/doi/10.1126/sciadv.abb5813
Genomes
Genomes
EPI_ISL_16743861, EPI_ISL_16743865, EPI_ISL_16975274, EPI_ISL_17078585-17078586, EPI_ISL_17094219, EPI_ISL_17094293, EPI_ISL_17146442, EPI_ISL_17180839, EPI_ISL_17181019, EPI_ISL_17182937, EPI_ISL_17190102, EPI_ISL_17191687, EPI_ISL_17198378, EPI_ISL_17206421, EPI_ISL_17207252-17207258, EPI_ISL_17207261, EPI_ISL_17207269, EPI_ISL_17229372, EPI_ISL_17229380, EPI_ISL_17236292, EPI_ISL_17236472, EPI_ISL_17236495, EPI_ISL_17236498, EPI_ISL_17237297, EPI_ISL_17238401, EPI_ISL_17238638, EPI_ISL_17240910The text was updated successfully, but these errors were encountered: