https://pymonnto.readthedocs.io/
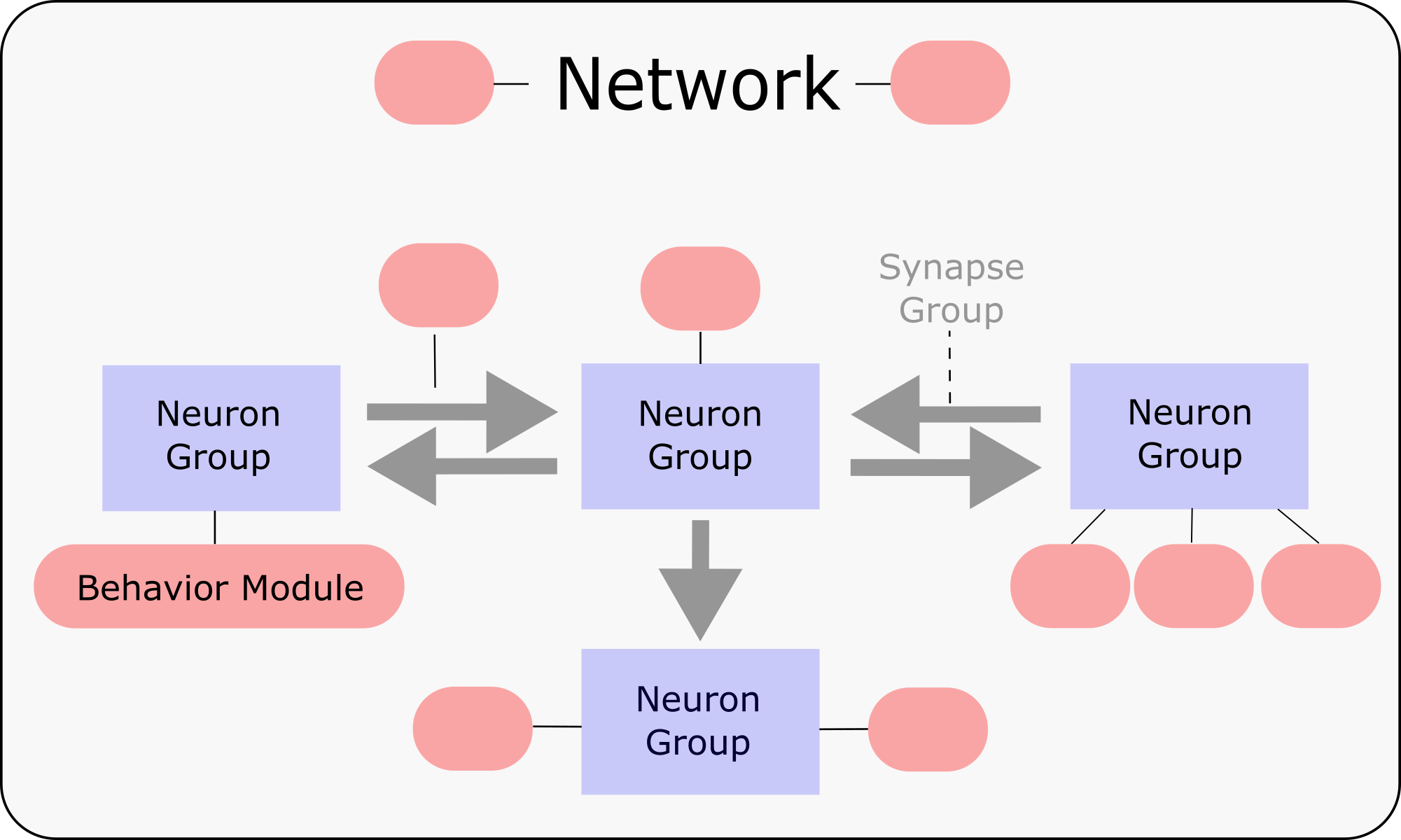
The "Python modular neural network toolbox" allows you to create different Neuron-Groups, define their Behavior and connect them with Synapse-Groups.
With this Simulator you can create all kinds of biological plausible networks, which, for example, mimic the learning mechanisms of cortical structures.
PymoNNto has many useful tools to speed up research and development, which are not limited to neural networks. Examples for this are the Evolution package for parameter optimization or the Storage Manager for data access.
PymoNNto requires Python3 and can be installen via pip with the following command:
pip install PymoNNto
If you want to extend the PymoNNto code you can also clone the git repository with
git clone [email protected]:trieschlab/PymoNNto.git --branch master --depth 1
and manually install the packages defined in requirements.txt
The following code creates a network of 100 neurons with recurrent connections and simulates them for 1000 iterations. What is still missing are some behavior modules. This modules have to be passed to the NeuronGrop to definde what the neurons are supposed to do at each timestep.
from PymoNNto import *
My_Network = Network()
NeuronGroup(net=My_Network, tag='my_neurons', size=100)
SynapseGroup(net=My_Network, src='my_neurons', dst='my_neurons', tag='GLUTAMATE')
My_Network.initialize()
My_Network.simulate_iterations(1000)Each Behavior Module has the following layout where initialize is called when the Network is initialized, while
iteration is called repeatedly every timestep. neurons points to the parent neuron group the behavior belongs to or some other parent object the beaviour is attached to.
In this example we define a variable voltage and a threshold. The activity-vector is initialized with 0 values for each neuron.
At each timestep a spike is created if the voltage is above the threshold value. After that, the voltage vector is decreased with factor 0.9 and random input is added.
When we combine the previous code blocks we can add the Basic_Behavior to the NeuronGroup.
The number in front of each behavior (1 and 9) have to be positive and determine the order of execution for each module inside and accross NeuronGroups.
To plot the neurons activity over time, we also have to create a Recorder. Here the voltage and the mean-voltage are stored at each timestep.
At the end the data is plotted via the tagging system (see tagging system section).
class Basic_Behavior(Behavior):
def initialize(self, neurons):
neurons.voltage = neurons.vector()
self.threshold = 0.5
def iteration(self, neurons):
neurons.spike = neurons.voltage > self.threshold
neurons.voltage[neurons.spike] = 0.0 #reset
neurons.voltage *= 0.9 #voltage decay
neurons.voltage += neurons.vector('uniform',density=0.01) #noise
...
# Add behavior to NeuronGroup
NeuronGroup(net=My_Network, tag='my_neurons', size=100, behavior={
1: Basic_Behavior(),
9: Recorder(['voltage', 'np.mean(voltage)'], tag='my_recorder')
})
...
import matplotlib.pyplot as plt
plt.plot(My_Network['n.voltage', 0, 'np'][:,0:10])
plt.plot(My_Network['np.mean(n.voltage)', 0], color='black')
plt.axhline(My_Network.my_neurons.Basic_Behavior.threshold, color='black')
plt.show()We can add more behavior modues to make the activity of the neurons more complex. Here the module Input_Behavior is added.
In initialize the synapse matrix is created, which stores one weight-value from each neuron to each neuron.
The Function iteration defines how the information is propagated to each neuron (dot product) and adds some term for random input.
The for loops are not neccessary here, because we only have one SynapseGroup.
This solution, however, also works with multiple Neuron- and SynapseGroups.
With synapse.src and synapse.dst you can access the source and destination NeuronGroups assigned to a SynapseGroup.
Here we also add an event recorder to plot the spikes of the NeuronGroup.
from PymoNNto import *
class Basic_Behavior(Behavior):
...
class Input_Behavior(Behavior):
def initialize(self, neurons):
for synapse in neurons.synapses(afferent, 'GLUTAMATE'):
synapse.W = synapse.matrix('uniform', density=0.1)
synapse.enabled = synapse.W > 0
def iteration(self, neurons):
for synapse in neurons.synapses(afferent, 'GLUTAMATE'):
neurons.voltage += synapse.W.dot(synapse.src.spike) / synapse.src.size * 10
My_Network = Network()
NeuronGroup(net=My_Network, tag='my_neurons', size=100, behavior={
1: Basic_Behavior(),
2: Input_Behavior(),
9: Recorder(['voltage', 'np.mean(voltage)'], tag='my_recorder'),
10: EventRecorder('spike', tag='my_event_recorder')
})
my_syn = SynapseGroup(net=My_Network, src='my_neurons', dst='my_neurons', tag='GLUTAMATE')
My_Network.initialize()
My_Network.simulate_iterations(1000)
# plotting:
import matplotlib.pyplot as plt
plt.plot(My_Network['voltage', 0, 'np'][:, 0:10])
plt.plot(My_Network['np.mean(voltage)', 0], color='black')
plt.axhline(My_Neurons['Basic_Behavior', 0].threshold, color='black')
plt.xlabel('iterations')
plt.ylabel('voltage')
plt.show()
plt.plot(My_Network['spike.t', 0], My_Network['spike.i', 0], '.k')
plt.xlabel('iterations')
plt.ylabel('neuron index')
plt.show()
To access the tagged objects we can use the [] operator which returns a list of objects tagged with the given tag.
E.g. ['my_tag'] gives you a list of all objects tagged with my_tag.
Tags searches use a caching system so it is very fast an can be used for repeated calls inside of Behaviors.
Unique tags can be accessed like simple variables via their parents like My_Network.my_neurons.
Here are some examples:
My_Network = Network()
NeuronGroup(net=My_Network, tag='my_neurons', size=100, behavior={
1: Basic_Behavior(),
9: Recorder(['voltage', 'np.mean(voltage)'], tag='my_recorder')
})
SynapseGroup(net=My_Network, src='my_neurons', dst='my_neurons', tag='S1,GLUTAMATE')
################################
################################
################################
#Unique tag access:
My_Network.my_neurons
My_Network.my_neurons.Basic_Behavior
My_Network.my_neurons.Recorder
My_Network.my_neurons.my_recorder
My_Network.S1
#[] tag search
My_Network['my_neurons']
=> [<PymoNNto.NetworkCore.Neuron_Group.NeuronGroup object at 0x00000195F4878670>]
My_Network['my_recorder']
My_Network.my_neurons['my_recorder']
=> [<PymoNNto.NetworkBehavior.Recorder.Recorder.Recorder object at 0x0000021F1B61D5E0>]
My_Network['np.mean(voltage)']
My_Network.my_neurons['voltage']
My_Network.my_neurons.my_recorder['np.mean(voltage)']
=> [[array(data iteration 1), array(data iteration 2), array(data iteration 3), ...]]
My_Neurons['voltage', 0]
#is equivalent to
My_Neurons['voltage'][0] #User Interface
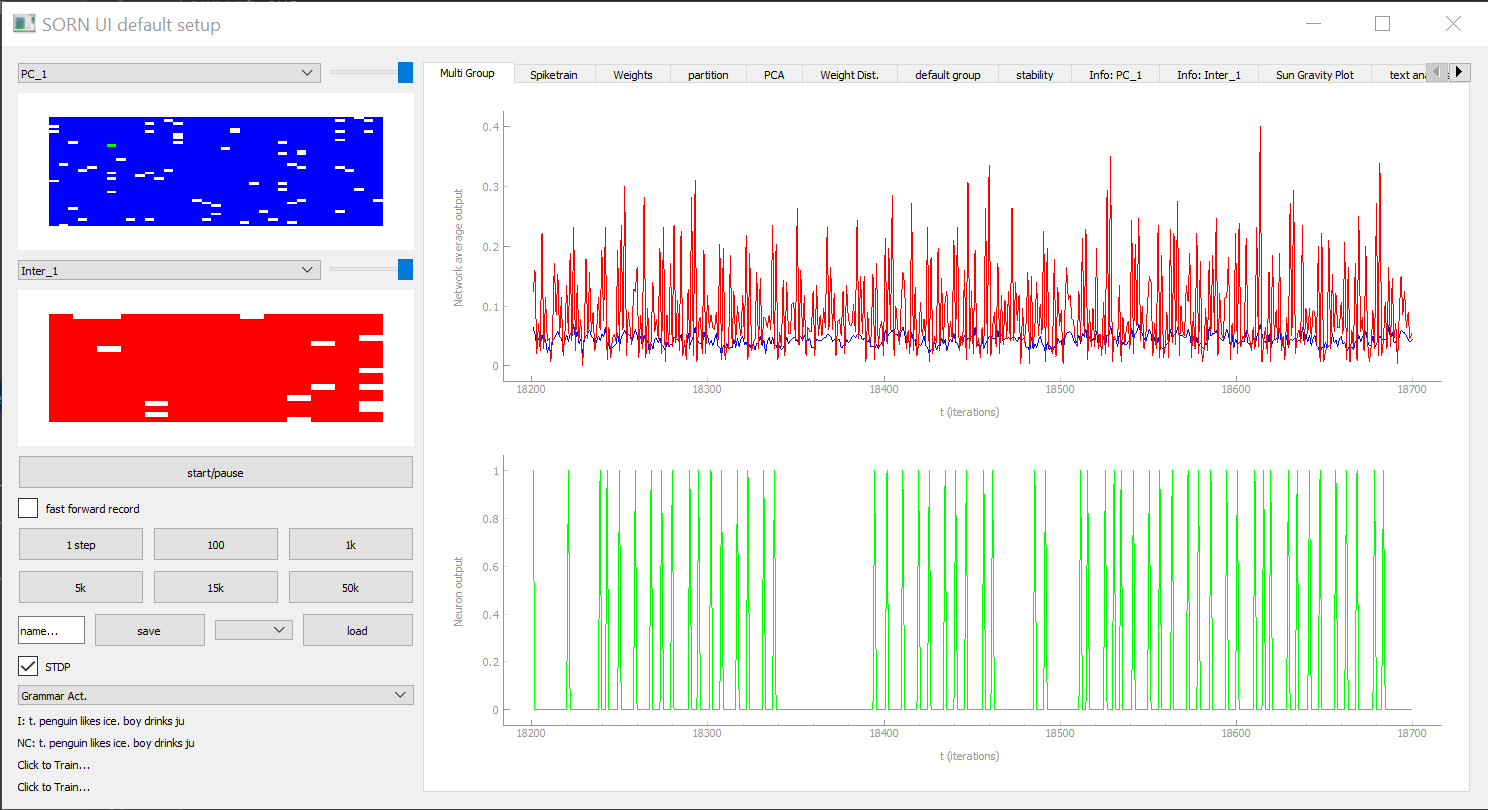
To control and evaluate the model from the "Basics" section with PymoNNto's interactive graphical user interface we can replace the pyplot functions, the recorder and the simulate_iterations with code to launch the Network_UI.
Like other parts of PymoNNto, the Network_UI is modular. It consists of multiple UI_modules, which we can be freely chosen. Here, we use the function get_default_UI_modules to get a list of standard modules applicable to most networks.
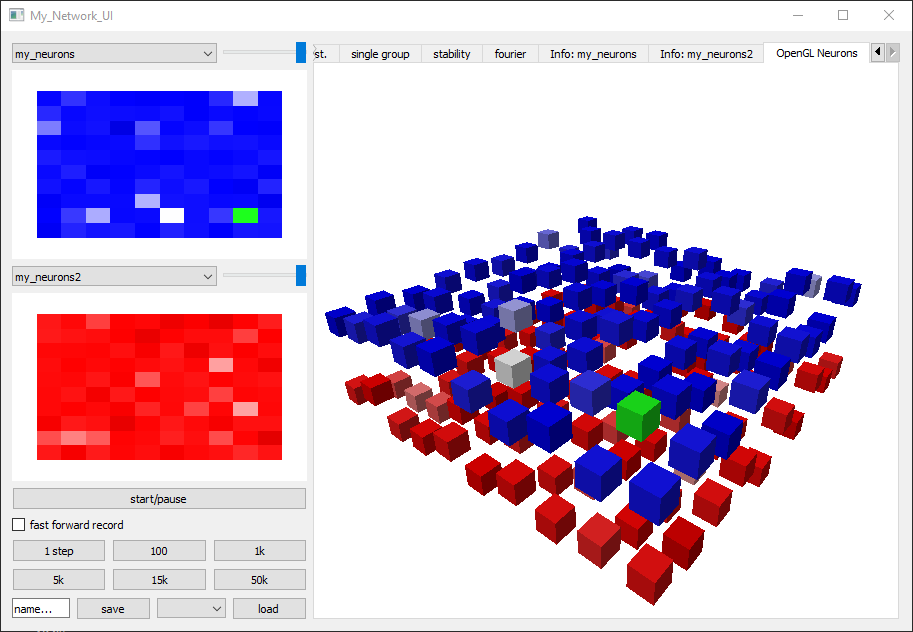
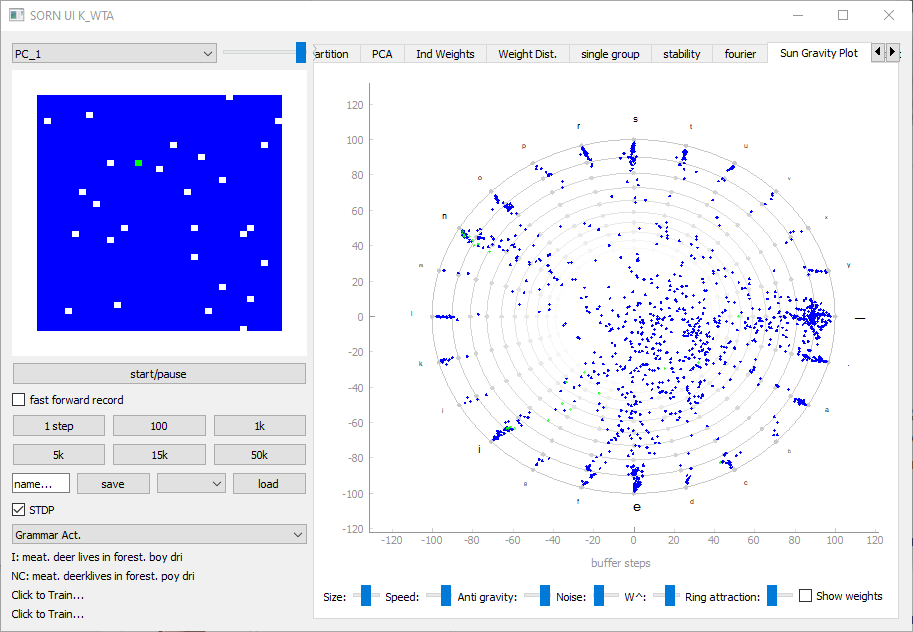
get_default_UI_modules receives a list of neuron variable names as well as a list of all the synapse variable names we want to display. The first neuron variable in the list will be used to set the brightness of the network activity grid module (blue rectangle on image)
Network_UI is initialized with the Network object, the ui module list, the window title. Additional to this, the attribute group_display_count defines how much activity grid modules we want to display, in case we want to see multiple NeuronGroups at once. The base color of the grid module can be defined by setting the MyNeuronGroup.color=(r,g,b,alpha) variable.
We can also set group_tags=[] and transmitters=[] (lists of strings) manually if we only want to display groups with certain tags or transmitters.
If we set reduced_layout to True we can remove the axis labels of the plots, so we get bigger plotting areas and a cleaner look.
To correctly render the output, some UI_modules require additional specifications or adjustment of the code. In this example, the sidebar_activity_module, displays the activity of the neurons on a grid and allows to select individual neurons. The size of the grid is specified as a property of the neuron group via a NeuronDimension module (here replaced with get_squared_dim(100) to get a 10x10 grid), which creates spatial coordinates for each neuron stored in the vectors x, y and z.
from PymoNNto import *
class Basic_Behavior(Behavior):
def initialize(self, neurons):
neurons.voltage = neurons.vector()
self.leak_factor = 0.9
self.threshold = 0.5
def iteration(self, neurons):
firing = neurons.voltage > self.threshold
neurons.spike = firing.astype(neurons.def_dtype) # spikes
neurons.voltage[firing] = 0.0 # reset
neurons.voltage *= self.leak_factor # voltage decay
neurons.voltage += neurons.vector('uniform', density=0.01) # noise
class Input_Behavior(Behavior):
def initialize(self, neurons):
for synapse in neurons.synapses(afferent, 'GLUTAMATE'):
synapse.W = synapse.matrix('uniform', density=0.1)
synapse.enabled = synapse.W > 0
def iteration(self, neurons):
for synapse in neurons.synapses(afferent, 'GLUTAMATE'):
neurons.voltage += synapse.W.dot(synapse.src.spike) / synapse.src.size * 10
My_Network = Network()
My_Neurons = NeuronGroup(net=My_Network, tag='my_neurons', size=get_squared_dim(100), behavior={
1: Basic_Behavior(),
2: Input_Behavior()
})
My_Neurons.visualize_module()
my_syn = SynapseGroup(net=My_Network, src='my_neurons', dst='my_neurons', tag='GLUTAMATE')
My_Network.initialize()
My_Network.simulate_iterations(1000, measure_block_time=True)
from PymoNNto.Exploration.Network_UI import *
my_UI_modules = get_default_UI_modules(['voltage', 'spike'], ['W'])
Network_UI(My_Network, modules=my_UI_modules, label='My_Network_UI', group_display_count=1).show()There is a huge variety of useful UI Tabs. Examples can be seen here:
This is an example how to implement an experiment with alternating training and testing stimuli.
trainging_stimulus = ...
testing_stimulus = ...
NeuronGroup(..., behavior={...,
1: InputStimulus(tag='train', stimulus=trainging_stimulus),
2: InputStimulus(tag='test', stimulus=testing_stimulus),
# ...
9: Recorder(tag="rec", variables=["n.v", "n.fired"]),
})
# initialize
for loop in range(loop_count):
network.activate_mechanisms('train')
network.deactivate_mechanisms('test')
network.train.index = 0 # [tag,0] is equivalent to [tag][0] and gives you the first found object with the given tag
network.simulate_iterations(len(trainging_stimulus))
recorded = network["n.fired", 0]
network.rec.reset()
network.deactivate_mechanisms('train')
network.activate_mechanisms('test')
network.test.index = 0
network.simulate_iterations(len(testing_stimulus))
recorded = network["n.v", 0]
network.rec.reset()
#increase the self.index variable inside of the InputStimulus module at each step.