Swinburne and Marinica, Phys. Rev. Lett 2018 (bibtex citation).
PAFI uses LAMMPS and mpi4py,
in addition to numpy scipy pandas. If you can run
from mpi4py import MPI
from lammps import lammps
lmp = lammps(comm=MPI.COMM_WORLD)
lmp.close()Then you can install PAFI:
pip install pafiTest routines can be found in this repository at tests/.
Otherwise, see here for installation on HPC clusters.
For local testing you can install LAMMPS via conda-lammps (one CPU/worker)
conda config --add channels conda-forge # add conda-forge channel
conda create -n pafi-env python=3.10
conda activate pafi-env
pip install numpy scipy pandas
conda install mpi4py lammps
pip install pafiWe emphasize conda-lammps will not give optimal performance on HPC!
See here for PAFI example scripts.
PAFI requires that you have already made some NEB calculation with some potential. You can then run
mpirun -np ${NUM_PROCS} python simple_pafi_run.pysimple_pafi_run.py:
from mpi4py import MPI
from pafi import PAFIManager, PAFIParser
parameters = PAFIParser()
parameters.set_potential(["neb_pathway/Fe.eam.fs"], # List of potential files
"eam/fs", # LAMMPS pair style
["Fe"]) # List of Elements
parameters.set_pathway("neb_pathway/image_*.dat") # NEB pathway of LAMMPS dat files
parameters.axes["Temperature"] = [100.*i for i in range(7)] # temperature range
parameters.set("CoresPerWorker",1) # Will force to 1 for conda installation of lammps
# Typical production values
parameters.set("SampleSteps",2000) # Sampling steps per worker
parameters.set("ThermSteps",1000) # Thermalization steps per worker
parameters.set("ThermWindow",500) # Averaging window to check temperature
parameters.set("nRepeats",1) # Number of times to repeat process (if CPU limited)
# Establish PAFIManager
manager = PAFIManager(MPI.COMM_WORLD,parameters=parameters)
manager.run()
manager.close()This will return a *.csv file with data readable by pandas. Can easily collate multiple runs.
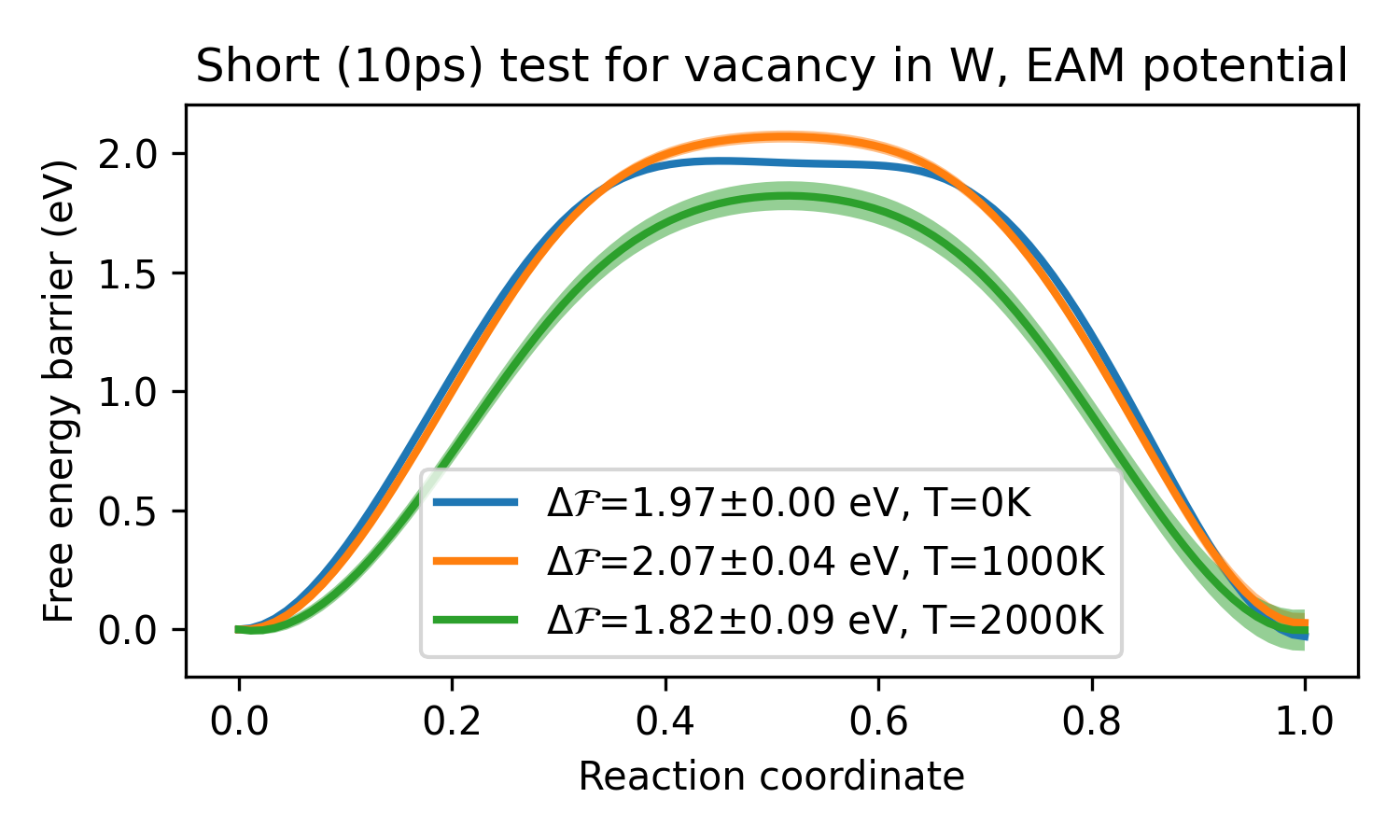
We can load in multiple csv files from some run then plot in python:
import matplotlib.pyplot as plt
from glob import glob
from pafi import ResultsProcessor
p = ResultsProcessor( data_path=glob("dumps/pafi_data_*.csv"),
xml_path='dumps/config_0.xml', integrate=True)
plotting_data,x_key,y_key = p.plotting_data()
# Plotting
fig = plt.figure(figsize=(5,3))
ax = fig.add_subplot(111)
ax.set_title("Short (10ps) test for vacancy in W, EAM potential")
for i,row in enumerate(plotting_data):
# Plotting data
x,y,e = row[x_key], row[y_key], row[y_key+"_err"]
T = int(row['Temperature'])
label = r"$\Delta\mathcal{F}$=%2.2f±%2.2f eV, T=%dK"% (y.max(),e.max(),T)
# make plots
ax.fill_between(x,y-e,y+e,facecolor=f'C{i}',alpha=0.5)
ax.plot(x,y,f'C{i}-',lw=2,label=label)
# save
ax.legend(loc='best')
ax.set_xlabel("Reaction coordinate")
ax.set_ylabel("Free energy barrier (eV)")See the examples and hints and tips for more information
-
See LAMMPS NEB for making a pathway
-
New
equalstyle gives optimal spacing for force integration- e.g.fix neb all neb 1.0 parallel equal -
Modify one of
examples/configuration_files/*_REAL.xmlto load in your pathway and potential:
from mpi4py import MPI
from pafi import PAFIManager
manager = PAFIManager(MPI.COMM_WORLD,"/path/to/config.xml")
manager.run()
manager.close()-
See the tutorial for information on the
pafi-path-testroutine -
In general, we want a reference pathway with dense discretisation where energy gradients are large
-
The current non-smoothed spline implementation can oscillate between very similar image configurations, as a result, there should be non-negligible displacement between images
-
If your path isn't loading, try setting
LogLammps=1inconfig.xmlto check for bugs inlog.lammps -
If
SampleStepsis too large workers will make thermally activated "jumps" to nearby paths in the hyperplane. This will return a warning messageReference path too unstable for sampling.and increase error. If this happens, decreaseSampleStepsand increasenRepeats -
When running on
NPROCScores, we requireNPROCS%CoresPerWorker==0, so we have an integer number of workers -
The total number of force calls per worker is
nPlanes * (ThermSteps+SampleSteps) * nRepeats, spatially parallelised by LAMMPS acrossCoresPerWorkercores for each worker. -
Each PAFI worker runs at the same speed as LAMMPS. Increasing
CoresPerWorkerwill typically decrease execution time but also reducenWorkersand increase error, as we have less samples. -
If you are core-limited, the
nRepeatsoption forces workers to perform multiple independent sampling runs on each plane. For example, with all other parameters fixed, running on 32 cores withnRepeats=3is equivalent to running on 3*32=96 cores withnRepeats=1, but the latter will finish in a third of the time.
For more details please see
Swinburne and Marinica, Unsupervised Calculation of Free Energy Barriers in Large Crystalline Systems, Phys. Rev. Lett., 2018 link
Citation:
@article{PhysRevLett.120.135503,
title = {Unsupervised Calculation of Free Energy Barriers in Large Crystalline Systems},
author = {Swinburne, Thomas D. and Marinica, Mihai-Cosmin},
journal = {Phys. Rev. Lett.},
volume = {120},
issue = {13},
pages = {135503},
numpages = {6},
year = {2018},
month = {Mar},
publisher = {American Physical Society},
doi = {10.1103/PhysRevLett.120.135503},
url = {https://link.aps.org/doi/10.1103/PhysRevLett.120.135503}
}Some use cases of PAFI:
- Allera et al., Activation entropy of dislocation glide, arXiv, 2024 link
- Nahavandian et al., From anti-Arrhenius to Arrhenius behavior in a dislocation-obstacle bypass: Atomistic Simulations and Theoretical Investigation, Computational Materials Science, 2024 link
- Namakian et al., Temperature dependence of generalized stacking fault free energy profiles and dissociation mechanisms of slip systems in Mg, Computational Materials Science, 2024 link
- Namakian et al., Temperature dependent stacking fault free energy profiles and partial dislocation separation in FCC Cu, Computational Materials Science, 2023 link
- Baima et al., Capabilities and limits of autoencoders for extracting collective variables in atomistic materials science, Physical Chemistry Chemical Physics, 2022 link
- Sato et al., Anharmonic effect on the thermally activated migration of {101̄2} twin interfaces in magnesium, Materials Research Letters, 2021 link
- Restart files from pathway deviations
- Smoothed spline interpolation for more general reference pathways