-
-
Notifications
You must be signed in to change notification settings - Fork 245
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Dirichlet mixture of multinomials example #18
Conversation
|
Check out this pull request on See visual diffs & provide feedback on Jupyter Notebooks. Powered by ReviewNB |
This looks great! So excited to have a guide to this, and one that supports prediction. |
|
Ooh nice @bsmith89 ! I'll review by tomorrow night 🥳 |
Only because I didn't know that function existed until right now 🙃
Can you point me to docs on how to modify an InferenceData object?
Input/Output cell 12 is a very superficial version of this already, no? Do you have ideas about deeper comparisons to plot? |
I believe there are two ways you could do this:
LMK if that won't work, and I will try to help. Seems like there ought to be a way of creating a new
I'm sorry, I don't. I was hoping you did! It seems like such a comparison is something that ArviZ should help with, but I don't know of an ArviZ plot function to use. Maybe |
Turns out there is! with model_marginalized:
ppc = pm.fast_sample_posterior_predictive(trace_marginalized, keep_size=True)
# Concatenate with InferenceData object
trace_marginalized = az.concat(trace_marginalized, az.from_dict(posterior_predictive=ppc))
Messed around with this a bit just now, and I can't really get much that seems worthwhile. For now I think I'll stick with the superficial example, but using the more complete InferenceData object as constructed above. |
Works for me. I'm not sure that ArviZ has the right capabilities for such PP checks, but it's likely I just don't know all that's in there. |
|
|
Maybe put an issue on the ArviZ GitHub issues page to point out this limitation? |
|
I'm not sure yet it's a limitation from ArviZ actually (see my comment for this cell on the NB). I'd bet this is doable with the |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
This is a really good start @bsmith89 🤩
Overall, I think this notebook lacks context about the problem and model. I left comments below, trying to adopt the point of view of a user who thinks this model is what she needs but is not sure yet -- feel free to ask if anything is unclear.
Once these comments are adressed it should be ready to merge 🎉
examples/mixture_models/dirichlet_mixture_of_multinomials.ipynb
Outdated
Show resolved
Hide resolved
examples/mixture_models/dirichlet_mixture_of_multinomials.ipynb
Outdated
Show resolved
Hide resolved
examples/mixture_models/dirichlet_mixture_of_multinomials.ipynb
Outdated
Show resolved
Hide resolved
examples/mixture_models/dirichlet_mixture_of_multinomials.ipynb
Outdated
Show resolved
Hide resolved
examples/mixture_models/dirichlet_mixture_of_multinomials.ipynb
Outdated
Show resolved
Hide resolved
examples/mixture_models/dirichlet_mixture_of_multinomials.ipynb
Outdated
Show resolved
Hide resolved
examples/mixture_models/dirichlet_mixture_of_multinomials.ipynb
Outdated
Show resolved
Hide resolved
| @@ -0,0 +1,748 @@ | |||
| { | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I'm positive you can use az.plot_ppc with the kind and coords kwargs here (see https://arviz-devs.github.io/arviz/api/generated/arviz.plot_ppc.html).
The nice is that you'll use xarray indexing instead of classic numpy indexing. i and j are the sample and category, right?
Reply via ReviewNB
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
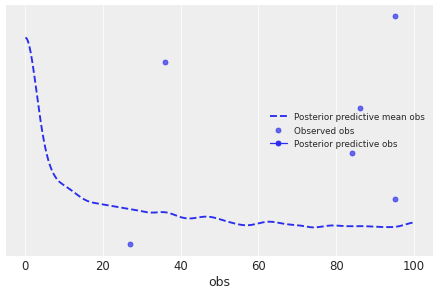
I can get plot_ppc to plot something using the coords= kwarg but it's not at all clear to me that it's set up well to produce a histogram like the one I made manually.
Here's what I tried:
az.plot_ppc(
trace_marginalized,
kind='scatter',
var_names=['obs'],
coords=dict(obs_dim_0=[9], obs_dim_1=[2])
)Having not read up on PPCs in ArviZ, it's not super apparent to me what that plot's trying to show. What's the y-axis? Frequency I assume, but then what are the 'o'-styled points? In any case, I'd much prefer to use a bar-chart style histogram than a KDE-plot, given that the values of obs are discrete integers.
Anyway, that's all to say that I don't think plot_ppc is currently the correct tool for this job.
Instead, to motivate the use of a PPC, I think I'll run a third model that's just a plain Multinomial distribution, and use the PPC to demonstrate that it does not fit the over-dispersed data well.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Interesting 🤔 Let me take a look ASAP, because I'm still not convinced that it's not currently possible with plot_ppc. Did you allow push by maintainers on this PR?
Also tagging @OriolAbril and @aloctavodia, who are ArviZ wizards 🧙♂️
I'll run a third model that's just a plain Multinomial distribution, and use the PPC to demonstrate that it does not fit the over-dispersed data well.
Great idea!
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I'm still not convinced that it's not currently possible with plot_ppc.
Looking forward to being proven wrong 😄
Did you allow push by maintainers on this PR?
Looks like that box is checked, yes.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Perfect, thanks. I'll tell you when I got some time to look over it
Looking forward to being proven wrong 😄
Ha ha, sorry, that's not what I meant -- rather, I'm seeing this as a nice, non trivial test case for plot_ppc's capacities 😉
Yep! That sounds about right. @ricardoV94 may also have some ideas for fleshing out context/motivation on this nb.
Great. Thanks for the detailed comments. I'll try and address them later this week. |
|
Hey @bsmith89 ! Did you get some time to make the changes? Any questions? |
|
I have not made changes yet, no. @ricardoV94 and I were discussing potentially making a more involved topic modeling example. You can see some of that discussion in this Discourse thread. It's been challenging to find a realistic example without running into problems during sampling. If the goal is to get this merged before the next release I think making the changes you have suggested to this existing example is probably the best approach. I'll take another look this evening or tomorrow. |
|
I see, thanks!
Well, we definitely want polished and well tought-out tutorial NBs, so that sounds awesome! And let's not rush it -- quality is more important than speed here. How much time do you guys think you need to write down this kind of example? Depending on this, we could merge the current simple tutorial (after making the changes I have suggested) to get something out with the next release, and replace it with the full case study when the latter is out? |
Yes, I think it will be challenging to identify a state-of-the-art topic-modeling example that doesn't run into sampling challenges (because most models are symmetrical to permutations of the topic labels, and therefore multimodal). Clearly we have dealt with that in other examples (e.g. the matrix factorization example, but I think it will require some thought about the best thing to do for our audience.
This is my vote. I think, with the changes you suggested, the current example will get users up-and-running. We can then extend it with a more realistic example of e.g. topic modeling after that. |
That makes total sense. Really looking forward to reviewing this -- hope I can be of help! And feel free to open questions on Discourse about this 😉
Yep, sounds good, let's do that then! Thanks in advance, and let me know when I can review the changes 🥳 |
|
Hey, sorry for the delay on this. I've updated the notebook with what I think might be the final form of the simulation/analysis for this first iteration of the example. This includes a "null", multinomial model of the data, PPCs comparing this to the marginalized model, and a final model comparison (LOO). What I haven't done is add any of the written explanations for the code. Instead I copied your comments, @AlexAndorra, into markdown cells as placeholders for the text that should be there. I think I can work on this for a small amount of time this week, hopefully getting it into merge-able form by Friday. However, if anyone (e.g. @ricardoV94) else would like to take a crack at writing the text, I'd be happy to relinquish that work. ;) I'm also happy to swap out the custom PPC plotting code for an ArviZ approach, if anyone has ideas for an effective figure. Let me know if you see more that should be changed besides the markdown cells. |
examples/mixture_models/dirichlet_mixture_of_multinomials.ipynb
Outdated
Show resolved
Hide resolved
examples/mixture_models/dirichlet_mixture_of_multinomials.ipynb
Outdated
Show resolved
Hide resolved
examples/mixture_models/dirichlet_mixture_of_multinomials.ipynb
Outdated
Show resolved
Hide resolved
|
I left some comments above. I think the NB is very interesting. It would be cool to have a clear posterior predictive sampling plot that shows how the Multinomial-only model generates underdispersed predictions relative to the observed data. I would add graphviz models to the 3 models, but that is a personal preference that others may not share. Overall, I think it is really nice and concise. PS: I don't know if I will have time to write the Markdown, so if you have free time feel free to jump in. |
|
I think I made all the changes we talked about, and this is ready for final review. Let me know what you think! |
examples/mixture_models/dirichlet_mixture_of_multinomials.ipynb
Outdated
Show resolved
Hide resolved
|
@bsmith89 I left some suggestions. Let me know if you have any questions :) |
|
I went through and made changes as suggested by you three, @AlexAndorra, @ricardoV94, and @MarcoGorelli . I then went ahead and "resolved" most of the inline conversations where I thought I had sufficiently addressed them. The only two things that I've left open are
@AlexAndorra, yours is the only unresolved code review. Once you're satisfied, I think this can be merged. |
| @@ -0,0 +1,1815 @@ | |||
| { | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Simplify to "Thankfully, we only get one warning related to inefficient sampling"?
Reply via ReviewNB
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Again, it's because Metropolis isn't as good as NUTS to spot and warn about problematic geometries in the posterior. I'd reformulate as: "we only get one warning now, so it seems that our sampling was more efficient".
Also, I think you should show how this model runs with NUTS, so that people can see what inefficient sampling looks like
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Also, I think you should show how this model runs with NUTS, so that people can see what inefficient sampling looks like
Unfortunately, NUTS fails entirely with:
---------------------------------------------------------------------------
RemoteTraceback Traceback (most recent call last)
RemoteTraceback:
"""
Traceback (most recent call last):
File "~/Projects/pymc3/pymc3/parallel_sampling.py", line 191, in _start_loop
point, stats = self._compute_point()
File "~/Projects/pymc3/pymc3/parallel_sampling.py", line 216, in _compute_point
point, stats = self._step_method.step(self._point)
File "~/Projects/pymc3/pymc3/step_methods/arraystep.py", line 276, in step
apoint, stats = self.astep(array)
File "~/Projects/pymc3/pymc3/step_methods/hmc/base_hmc.py", line 159, in astep
raise SamplingError("Bad initial energy")
pymc3.exceptions.SamplingError: Bad initial energy
"""
The above exception was the direct cause of the following exception:
SamplingError Traceback (most recent call last)
SamplingError: Bad initial energy
The above exception was the direct cause of the following exception:
ParallelSamplingError Traceback (most recent call last)
<ipython-input-22-16dea9c651bc> in <module>
1 with model_multinomial:
----> 2 trace_multinomial = pm.sample(
3 draws=int(5e3), chains=4, return_inferencedata=True
4 )
~/Projects/pymc3/pymc3/sampling.py in sample(draws, step, init, n_init, start, trace, chain_idx, chains, cores, tune, progressbar, model, random_seed, discard_tuned_samples, compute_convergence_checks, callback, jitter_max_retries, return_inferencedata, idata_kwargs, mp_ctx, pickle_backend, **kwargs)
555 _print_step_hierarchy(step)
556 try:
--> 557 trace = _mp_sample(**sample_args, **parallel_args)
558 except pickle.PickleError:
559 _log.warning("Could not pickle model, sampling singlethreaded.")
~/Projects/pymc3/pymc3/sampling.py in _mp_sample(draws, tune, step, chains, cores, chain, random_seed, start, progressbar, trace, model, callback, discard_tuned_samples, mp_ctx, pickle_backend, **kwargs)
1473 try:
1474 with sampler:
-> 1475 for draw in sampler:
1476 trace = traces[draw.chain - chain]
1477 if trace.supports_sampler_stats and draw.stats is not None:
~/Projects/pymc3/pymc3/parallel_sampling.py in iter(self)
477
478 while self._active:
--> 479 draw = ProcessAdapter.recv_draw(self._active)
480 proc, is_last, draw, tuning, stats, warns = draw
481 self._total_draws += 1
~/Projects/pymc3/pymc3/parallel_sampling.py in recv_draw(processes, timeout)
357 else:
358 error = RuntimeError("Chain %s failed." % proc.chain)
--> 359 raise error from old_error
360 elif msg[0] == "writing_done":
361 proc._readable = True
ParallelSamplingError: Bad initial energy
I messed around with target_accept and testval, but I couldn't get all 4-chains to finish without numerical issues.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Copying my comments here because they're relevant to this discussion as well:
I haven't drilled down into the NUTS sampling errors, but I'm guessing that we're getting logp's or dlogp's of-infdue to the large deviation from the multinomial predictive density (e.g. maybeobserved_counts[0, 4] == 4is just too unbelievable?).
Since Metropolis either (A) rejects jumps into regions of the parameter space where logp is -inf, or (B) doesn't know about the numerically unstable gradient, it doesn't fail the same way NUTS does.
| @@ -0,0 +1,1815 @@ | |||
| { | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
|
@bsmith89 I think the last changes are really great. I left two minor sentence suggestions. More importantly I replied to our thread on the "extra" parameter of the DM. Let me know if I explained myself well. |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
This is shaping up to be a really good example NB, I really love what you did here @bsmith89 🤩
There is some context about the usefulness of DM at the beginning, and mention of its shortcomings at the end (and you don't just say "hey you can use something else than DM when that happens", you actually point to alternatives 👏 ).
I left a few comments and suggestions below. This should polish the NB even more, and once that is done I think we can merge 🥳
examples/mixture_models/dirichlet_mixture_of_multinomials.ipynb
Outdated
Show resolved
Hide resolved
examples/mixture_models/dirichlet_mixture_of_multinomials.ipynb
Outdated
Show resolved
Hide resolved
examples/mixture_models/dirichlet_mixture_of_multinomials.ipynb
Outdated
Show resolved
Hide resolved
examples/mixture_models/dirichlet_mixture_of_multinomials.ipynb
Outdated
Show resolved
Hide resolved
| @@ -0,0 +1,1815 @@ | |||
| { | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
- By n, you mean n_samples here, right? If yes, would be better to use this denomination, to match the one in the code
- "While either of these would probably overcome the problems with the default sampler": why is so?
- "here we'll instead switch to a standard, Metropolis step method, increasing our number of draws to overcome the inefficiency relative to NUTS": this formulation is problematic to me, as the problem doesn't come from NUTS, it comes from the model. Switching to Metropolis doesn't make the model more efficient; it just makes the warnings go away, both because Metropolis just doesn't have them, and because the algorithm itselft is less robust. The current framing implies that switching to Metropolis makes the convergence problems go away when it just hides them. As it's a mistake that's often made by people coming to Bayes, I think we should change the formulation above. What do you think?
Reply via ReviewNB
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I haven't drilled down into the NUTS sampling errors, but I'm guessing that we're getting logp's or dlogp's of -inf due to the large deviation from the multinomial predictive density (e.g. observed_counts[3, 1] == 1 is pretty improbable given that the expectation is 15).
Switching to Metropolis doesn't make the model more efficient; it just makes the warnings go away, both because Metropolis just doesn't have them, and because the algorithm itselft is less robust. The current framing implies that switching to Metropolis makes the convergence problems go away when it just hides them.
Since Metropolis either (A) rejects jumps into regions of the parameter space where logp is -inf, or (B) doesn't even know about the numerically unstable gradient, it doesn't fail the way NUTS does.
"While either of these would probably overcome the problems with the default sampler"
Haven't actually tried it, but I'm conjecturing here that the Bernoulli categorical likelihood elements (note: Bernoulli was a mistake) won't be as "surprised" (so to say) with hits to the low frequency species since any one observation is fairly believable. Equivalently with the collapsed likelihood, since there's nothing surprising about any combination of species counts. (...Does this explanation make any sense?)
(Addendum: I went ahead and tried out the collapsed model, which works as expected.)
this formulation is problematic to me [...] As it's a mistake that's often made by people coming to Bayes, I think we should change the formulation above.
Do you have a suggestion for better wording?
What do you think?
Mmm...I'm pretty okay with mostly sweeping these issues under the rug in this example. I think the details you're considering here should be in an e.g. "Diagnosing Biased Inference" or "Reparameterizing Problematic Models" notebooks. Here that problem serves as good motivation for using a better model, but I don't think we need to deal with it much more than that.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I agree that it might be a bit out of the scope of this Notebook to explore why NUTS fails. Maybe just warn more explicitly that this is not a real solution to the problem, but just a shortcut so you have something to compare with the proper DM model?
And later don't emphasize so much that the diagnostics look nice, but caution again that while they may look nicer we are probably glossing over very real problems in the posterior approximation that the NUTS made clear.
(All this is in line with the Folk theorem right? We are seeing evidence of a misspecified model)
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I'm guessing that we're getting logp's or dlogp's of-infdue to the large deviation from the multinomial predictive density (e.g.observed_counts[3, 1] == 1is pretty improbable given that the expectation is 15).
Yeah, that'd be my guess too. We could probably help NUTS run by selecting better priors than the current uniform ones, but, by design, the issue is deeper -- the model is just not a good enough approximation of the generative process.
Does this explanation make any sense?
Yeah thanks, I think I understand what you mean. You can see if you can briefly add that in the text above, but it's not a blocker.
'm pretty okay with mostly sweeping these issues under the rug in this example. [...] Here that problem serves as good motivation for using a better model, but I don't think we need to deal with it much more than that.
I agree with that. What I'm saying is we should say that doing so is not fixing issues but sweeping them under the rug, otherwise readers could think that just switching to Metropolis when NUTS breaks is a valid workflow, whereas the latter happening should be a red alert -- actually, I think there'd be value in mentioning or even showing that NUTS doesn't even want to sample such a model --> folk theorem must pop in our head.
Do you have a suggestion for better wording?
The way you reworded it is already quite good! I'd just add a bit to it. Something like:
"... here we'll instead switch to the Metropolis step method, which ignores some of the geometric pathologies of our naïve model. This is important: switching to Metropolis does not not fix our model's issues, rather it sweeps them under the rug. In fact, if you try running this model with PyMC's default sampler, NUTS will break loudly from the beginning. When that happens, this should be a red alert that there is something wrong in our model.
You'll also notice below that we have to increase considerably the number of draws we take from the posterior, because Metropolis is much less efficient than NUTS."
Ricardo's last comment is also spot-on:
And later don't emphasize so much that the diagnostics look nice, but caution again that while they may look nicer we are probably glossing over very real problems in the posterior approximation that the NUTS made clear.
examples/mixture_models/dirichlet_mixture_of_multinomials.ipynb
Outdated
Show resolved
Hide resolved
examples/mixture_models/dirichlet_mixture_of_multinomials.ipynb
Outdated
Show resolved
Hide resolved
examples/mixture_models/dirichlet_mixture_of_multinomials.ipynb
Outdated
Show resolved
Hide resolved
examples/mixture_models/dirichlet_mixture_of_multinomials.ipynb
Outdated
Show resolved
Hide resolved
| @@ -0,0 +1,1815 @@ | |||
| { | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Great plot
Thanks! Just fancy-fied it even more with KDEs of the posterior samples and legends.
Thoughts? Too busy? Should the discussion/visualization of biased parameter estimation be split out entirely to a forest plot?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I think, as you said, it's now better to remove the posterior and true. The forest plots illustrates it perfectly, and this one looks a bit too busy with everything in it. The new legend works nicely btw.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I wrote that above: I think the posterior can be removed but the true values don't hurt as those are just lines
|
View / edit / reply to this conversation on ReviewNB ricardoV94 commented on 2021-01-25T08:36:33Z Cool! AlexAndorra commented on 2021-01-25T10:38:46Z This is great, thanks for adding it! Just one comment: you need to call the ArviZ function directly: |
|
This is great, thanks for adding it! Just one comment: you need to call the ArviZ function directly: View entire conversation on ReviewNB |
|
View / edit / reply to this conversation on ReviewNB AlexAndorra commented on 2021-01-25T10:56:03Z Looks neat now! Just two comments:
|
|
View / edit / reply to this conversation on ReviewNB AlexAndorra commented on 2021-01-25T10:56:03Z Nice! The legend is a bit cluttered now ha ha. I think you can remove the posterior from the plot and legend, as it's less core to this plot. You can also make the figure a bit wider |
|
View / edit / reply to this conversation on ReviewNB AlexAndorra commented on 2021-01-25T10:56:04Z "from the expected frequencies, |
|
View / edit / reply to this conversation on ReviewNB AlexAndorra commented on 2021-01-25T10:56:05Z Same here: use ArviZ directly |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Thanks a lot for the revisions @bsmith89 ! I answered to a few of your comments and left another few -- we're getting there 💪
|
I'm hoping that's everything. 🤞 ! |
Looks really great! Nice work. I have no more suggestions :). It's up to our editor @AlexAndorra now |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Yes, all good now 🥳 Thanks a lot for sticking to it @bsmith89, this is a great example!
Add an example to demonstrate the forthcoming DM distribution (see PyMC3 PR #4373).