-
Notifications
You must be signed in to change notification settings - Fork 136
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Usage questions about nglview: Please ask here first [1] #785
Comments
|
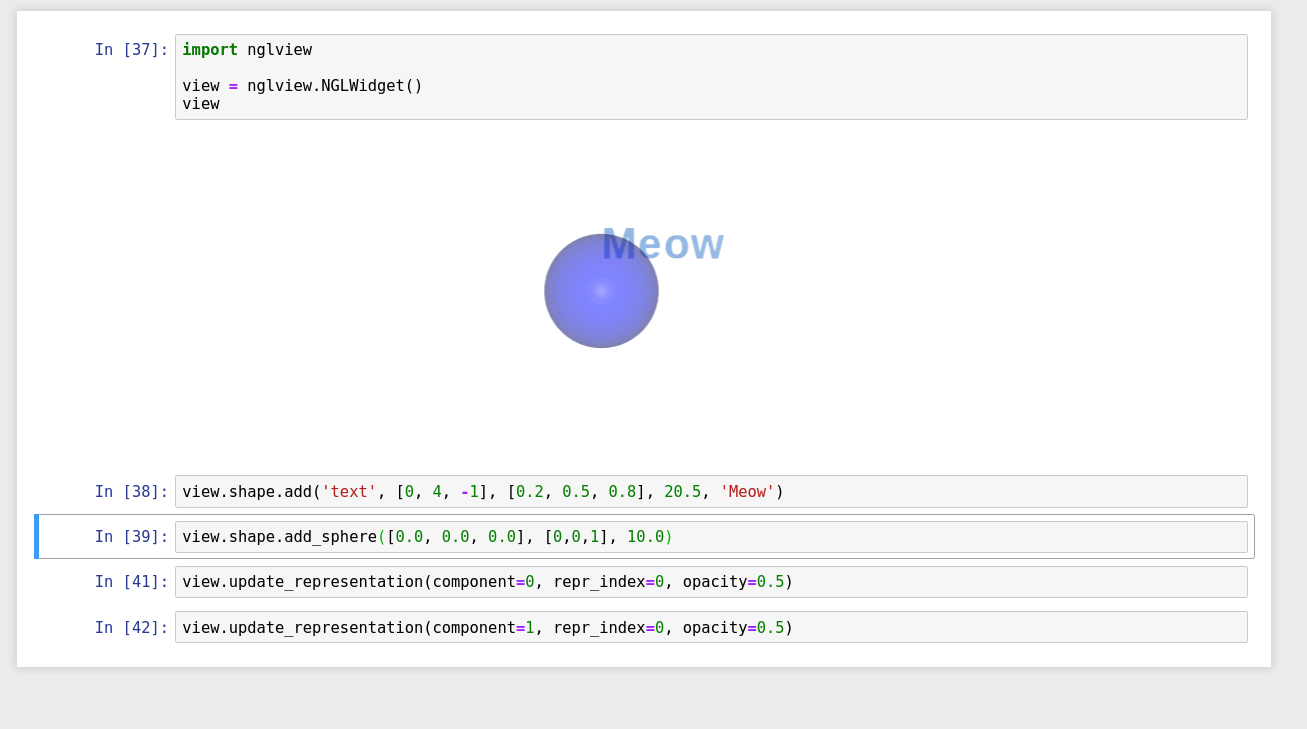
If I create a sphere with eg: view = nglview.NGLWidget()
view.shape.add_sphere([0.0, 0.0, 0.0], [0,0,1], 10.0)is there a way of setting the opacity of the sphere? Thanks in advance! |
|
hi, there's no API yet. But you can try with below code (using the latest version). |
Great, thanks! |
|
I'm trying to figure out the best way to set the orientation. Here is my current procedure. It works. However, can you let me know if this is indeed the best way. The reason that I wasn't sure is that it uses two private methods:
|
|
Hi,
It’s currently the best way. Cheers
Hai
…On Sun, Apr 28, 2019 at 3:38 PM Jesse Bloom ***@***.***> wrote:
I'm trying to figure out the best way to set the orientation. Here is my
current procedure. It works. However, can you let me know if this is indeed
the best way. The reason that I wasn't sure is that it uses two private
methods:
1.
Manually set the orientation to whatever I want by rotating the object.
2.
Get that orientation (as a 16-number list) with:
mat = view._camera_orientation
3.
In the future, set that orientation with:
view._set_camera_orientation(mat)
4.
If I want to make sure it is also centered, call:
view.center()
—
You are receiving this because you authored the thread.
Reply to this email directly, view it on GitHub
<#785 (comment)>, or mute
the thread
<https://github.com/notifications/unsubscribe-auth/ABB645J4BKIFR75FYLHPWUDPSX4LNANCNFSM4HHSP2TA>
.
|
@jbloom I've checked and we should use |
|
Hi everyone, The version of packages is as follows: |
|
I also had @mktonycho's problem with vertical lines, although they go away if I use |
|
Hi
Yeah, that is the current solution. I will open an issue in NGL. Thanks.
…On Tue, May 14, 2019 at 8:43 AM Jesse Bloom ***@***.***> wrote:
I also had @mktonycho <https://github.com/mktonycho>'s problem with
vertical lines, although they go away if I use antialias=True with
render_image.
—
You are receiving this because you authored the thread.
Reply to this email directly, view it on GitHub
<#785>,
or mute the thread
<https://github.com/notifications/unsubscribe-auth/ABB645KUL5HCGCSRUTVV3L3PVKXWFANCNFSM4HHSP2TA>
.
|
@mktonycho should be fixed in master branch (by upgrading to NGL 0.36). |
|
Hi! Not sure if this should be submitted as a separate issue, or if there is something I'm not doing correctly that can easily be fixed on my side. I'm trying to run nglview in a Jupyter Notebook through ASE using nglview.show_ase(atoms) where atoms is the ASE Atoms-object I want to view. When I make this call, I get a permission error is thrown (pasted below). Appreciate any help I can get here. PermissionError Traceback (most recent call last) C:\Anaconda3\lib\site-packages\nglview\show.py in show_ase(ase_atoms, **kwargs) C:\Anaconda3\lib\site-packages\nglview\widget.py in init(self, structure, representations, parameters, **kwargs) C:\Anaconda3\lib\site-packages\nglview\widget.py in add_structure(self, structure, **kwargs) C:\Anaconda3\lib\site-packages\nglview\widget.py in _load_data(self, obj, **kwargs) C:\Anaconda3\lib\site-packages\nglview\adaptor.py in get_structure_string(self) C:\Anaconda3\lib\site-packages\nglview\adaptor.py in _get_structure_string(write_method, suffix) C:\Anaconda3\lib\site-packages\ase\atoms.py in write(self, filename, format, **kwargs) C:\Anaconda3\lib\site-packages\ase\io\formats.py in write(filename, images, format, parallel, append, **kwargs) C:\Anaconda3\lib\site-packages\ase\io\formats.py in _write(filename, fd, format, io, images, parallel, append, **kwargs) C:\Anaconda3\lib\site-packages\ase\io\formats.py in open_with_compression(filename, mode) PermissionError: [Errno 13] Permission denied: 'C:\cygwin64\tmp\tmp7fatov62.pdb' |
|
hi @rasmusthog, you can do either
something like |
|
Hi, I'm trying to get nglview up and running and I'm probably overlooking something simple... I created an environment with conda and installed juypter, pytraj and nglview. Everything installed smoothly. When I run the tutorial, I see the expected output from the first cell (nglview version = 1.1.7, pytraj version = 2.0.4) However, after the third cell, (view._display_image()), I get a broken image icon. I tried running the same command sequence in ipython and got <IPython.core.display.Image object> as the output, but again, no image. I've tried with both python 2.7 and 3.7, both locally and remotely, and with the tutorial files and my own and I get the same behavior in all scenarios. The notebook is running on ubuntu 18.04. Thanks for your help |
|
Thanks @hainm. Here's the screenshot. I tried both with render_image and _display_image in the different and the same cells. I get the same behavior when I run the tutorial notebook in place or if I copy the commands to a new notebook and run it there. I tried updating to 2.1.0 and I still don't see the image, although now it's trying to display after the render_image step as you said. |
|
sorry for your inconvenience. Can you try
|
|
That worked! It looks great now. Thanks so much for the prompt help! |
|
thanks for trying. |
|
Thanks, @hainm! I have now modified the permissions (changed the ownership of the tmp-folder to my user instead of the local Administrator account), but I'm still getting the same error. Also tried changing the permissions so that the tmp-folder is globally readable (chmod -R 777), but still the same result. This is how the folder looks from a ls -l: drwxrwxrwx+ 1 [my username] [computer name]+None 0 May 30 10:27 tmp |
|
sorry @rasmusthog that I can't not help further with the permission error because I don't have any windows machine to test (I am mainly working with linux and macos). But please try the patch I mentioned. cheers. |
|
I'm embedding NGL. I'm loading a model like this (in an async typescript function): but when I dump the Three.js scene (stage.viewer.scene) immediately after this, the graphics objects don't exist yet. I think they're getting created in the background. |
|
@garyo please see example(s) here: https://github.com/arose/ngl/blob/master/examples/scripts/representation/distance.js |
|
Thanks for the example -- but I don't see there how to invoke a callback once all the graphics objects are loaded. If you dump the scene at the end of that example, just after |
|
Looking at the source, in representation.ts |
|
Hello! Thanks for the great tool! I have a question about the jupyter notebook to html conversion. The images didn't show in the exported html. Is there a way to display the images within the whole notebook as a html? Thanks in advance !!😄 |
hi @wangfan860 unfortunately No. You can use |
|
I'm having a problem loading a .gsd and a .dcd file though MDAnalysis and visualizing with nglviewer. No matter what representation I use with the Widget object, I cannot get rid of the bonds between the atoms in my system (there should be none). My MDAnalysis Universe has no bonds, but the Widget shows connections between all of the atoms. I'm also having trouble based on the naming conventions of my system (it's course grained, so all atom names start with '_'), and cannot change the color of each of the two atom types individually. If anyone knows a good way to overwrite my atom names to something more easily recognized by nglview, that would be great. import nglview as nv
import MDAnalysis as mda
U = mda.Universe( 'bulk.gsd', 'trajectory_bulk_chi0.dcd')
for atom in U.atoms:
if atom.name == 'cg_A':
atom.residue.resname = 'RES'
atom.name = 'A'
else:
atom.residue.resname = 'RES'
atom.name = 'B'
print(U.bonds)
view = nv.show_mdanalysis(U)
view.add_representation('ball_and_stick', selection='.A', color='red')
view.add_representation('ball_and_stick', selection='.B', color='blue')
viewpython=3.7 . I have numeric residue names and I think it's causing problems using masks. Is there a way around this? python version: 3.9 Thanks, |
hi @markperri: Is your question about |
|
hi @hainm, oh you're right, sorry for the mixup! |
|
you can watch this thread: Amber-MD/cpptraj#930 |
|
Hi - I was wondering if there is an easy way of changing the element in a pre-existing view - i.e. to interactively change elements, without having to remake the entire widget? |
What do you mean about "elements"? Are they "components" in |
|
I guess you could say components. More specifically, I'm adding/removing individual atoms from an ASE trajectory, starting from 1 full atoms object, and then replacing single sites. I guess one option is to draw each atom in the entire atoms object as 1 component, and then add/remove them individually like that. |
Oh, I don't think |
|
Well - slightly on extension of this, how would you force an update? Say I make some changes to my components, and then do a |
|
@atygesen Please see this: #589 (comment) |
and here: |
|
Hi! |
dear @YoelShoshan, Assume you have two components in your view Lines 842 to 847 in e2f7fe5
|
|
@hainm Thank you very much! Please see the following small code reproducing my issue: is there a certain way I should wrap/convert the structure instance that I get from MMCIFParser for nglview to recognize it? |
Is that |
|
@hainm works perfectly, thanks! |
Another question - what are reasonable values for asl1 and asl2 ?
What are reasonable values to use and what is the format ? |
I think they use NGL's selection language here. Please have a look: https://nglviewer.org/ngl/api/manual/usage/selection-language.html |
|
Thanks everyone and @arose |
Hello, I am trying to view a mixed solvent system using the above code, but the lithium ions in this system do not seem to display properly, how should I solve this problem |


For any questions relating to how/why/ ... Or anything you don't know where to ask.
For bug, suggestion, ... please open seperate issue
PS: QA-0: #589
The text was updated successfully, but these errors were encountered: