Computational chemistry at cloud scale! ChemCloud Server exposes the BigChem distributed compute system via API endpoints. Interactive API documentation here
- qcio - Elegant and intuitive data structures for quantum chemistry, featuring seamless Jupyter Notebook visualizations.
- qcparse - A library for efficient parsing of quantum chemistry data into structured
qcioobjects. - qcop - A package for operating quantum chemistry programs using
qciostandardized data structures. Compatible withTeraChem,psi4,QChem,NWChem,ORCA,Molpro,geomeTRIC, and many more, featuring seamless Jupyter Notebook visualizations. - BigChem - A distributed application for running quantum chemistry calculations at scale across clusters of computers or the cloud. Bring multi-node scaling to your favorite quantum chemistry program, featuring seamless Jupyter Notebook visualizations.
ChemCloud- A web application and associated Python client for exposing a BigChem cluster securely over the internet, featuring seamless Jupyter Notebook visualizations.
poetry installpoetry run pre-commit install # installs hooks for commit stage
poetry run pre-commit install --hook-type pre-push # install hooks for push stageRun the ChemCloud Server alone with no BigChem compute backend. Go to http://localhost:8000/docs to view interactive documentation.
poetry run uvicorn chemcloud_server.main:app --reloadCheck that your installation is working correctly by running the tests. If tests fail see note below about allocating enough memory for docker.
Running tests uses the docker-compose.yaml service specification. To use docker-compose you must place a .env files in the root directory. You only need to do this once.
touch .envbash scripts/tests.shA test summary will be output to /htmlcov. Open /htmlcov/index.html to get a visual representation of the test coverage. bash scripts/tests.sh automatically stops all docker containers after running the tests.
Run the ChemCloud server and BigChem compute backend (rabbitmq, redis, and psi4-powered worker instance). The following will build images for the web server and pull down the latest BigChem worker image. It will mount the local code into the web server so that it hot-reloads any changes made to the codebase. The worker can actively pickup tasks and run them. Authentication will not work until the correct environment variables are added to the .env file, see Manage environment and Auth0 for local development below.
Start ChemCloud-server and BigChem compute backend (NOTE: on older machines the docker compose command may be docker-compose, with a hyphen -).
docker compose up -d --buildTo shutdown the application:
docker compose downFor more granularity you can use docker to run various components of the service and run components outside of docker. A good development setup is to run the BigChem backend fully dockerized, then run the ChemCloud server in your local environment for easier control as you develop.
Start BigChem backend
docker compose up -d bigchem-workerRun ChemCloud server on your local machine; it will connect automatically to the BigChem
poetry run uvicorn chemcloud_server.main:app --reloadStart desired services found in docker-compose.yaml
docker compose up -d --build [services-of-interest]Stop all services
docker compose downSettings are managed in chemcloud_server/config and the Settings object will automatically look for environment variables found in both the environment and a .env file. To enable authentication for local development add the following variables to a .env file in the root directory with their corresponding values. These values are not required for tests to run correctly. Outside of testing, authentication-protected endpoints (compute endpoints) will not work without auth setup. Set up an account on Auth0 and supply the required environment variables below and you'll have a fully functioning application with secure auth! On Auth0 you should setup an [API(https://auth0.com/docs/get-started/apis) and register with it a Regular Web Application as your app type. The API needs to have compute:public and compute:private permissions (scopes) created and assigned to users who perform computations. ChemCloud is currently configured to only check for the compute:public scope.
AUTH0_DOMAIN=
AUTH0_CLIENT_ID=
AUTH0_CLIENT_SECRET=
AUTH0_API_AUDIENCE=
If you want to run the application without auth, comment out the line in main.py containing: dependencies=[Security(bearer_auth, scopes=["compute:public"])]. THIS IS STRONGLY NOT RECOMMENDED, ESPECIALLY IF SERVING YOUR CHEMCLOUD APPLICATION ON THE OPEN INTERNET! SECURITY AND AUTHENTICATION ARE PARAMOUNT!
You may need 3GB+ of memory allocated to Docker in order for the tests to run correctly. Psi4, the quantum chemistry package used for running tests, requests memory resources that may exceed what Docker can provide if restricted to only 2GB of memory (the default setting). Insufficient memory will result in failing tests. Click Docker -> Preferences -> Resources to allocate more memory to your local docker engine. At least 4GB is recommended.
Developing on a machine with GPUs means you can run a BigChem worker containing TeraChem. Add docker/terachem.yaml to the docker compose command to run a BigChem worker with Terachem.
docker compose -f docker-compose.yaml -f docker/terachem.yaml up -d --buildFirst either set the path to your TeraChem license in the .env as shown below or run TeraChem in unlicensed mode by commenting out ${TERACHEM_LICENSE_PATH} in docker-compose.yaml. Not taking one of these two actions will result in TeraChem entering an infinite loop looking for a license without notifying the end user. The TeraChem server will appear to be operational but will not respond to requests.
In the .env file add:
TERACHEM_LICENSE_PATH=/path/to/terachem/license/on/local/machine.keyFull CI/CD is handled via CircleCi. See .circleci/config.yml for details.
In the directory on the server from which the docker-compose.web.yaml file will deploy, create a server.env file and populate it with the following secrets:
BASE_URL=https://yourdomain.com
BIGCHEM_BROKER_URL=amqp://${USERNAME}:${PASSWORD}@${DOCKER_SERVICE_NAME_FOR_BIGCHEM_BROKER}:5672 # pragma: allowlist secret
BIGCHEM_BACKEND_URL=redis://:${PASSWORD}@${DOCKER_SERVICE_NAME_FOR_BIGCHEM_BACKEND}:6379/0
AUTH0_DOMAIN=xxx
AUTH0_CLIENT_ID=xxx
AUTH0_CLIENT_SECRET=xxx # pragma: allowlist secret
AUTH0_API_AUDIENCE=xxxNote that the BigChem URLs should correspond to the internal docker service name for BigChem broker and backend, not the public URLs used by external BigChem workers. We want the web server to connect locally, rather than over the open internet, to these services.
For serving ChemCloud over the open internet, is recommended that you run it behind a traefik reverse proxy that provides TLS termination and all the other security features expected of a reverse proxy. The docker/docker-compose.web.yaml file demonstrates the configuration required to securely serve ChemCloud behind traefik. A good overview can be found here.
Visualize all your results with a single line of code!
First install the visualization module:
pip install qcio[view]or if your shell requires '' around arguments with brackets:
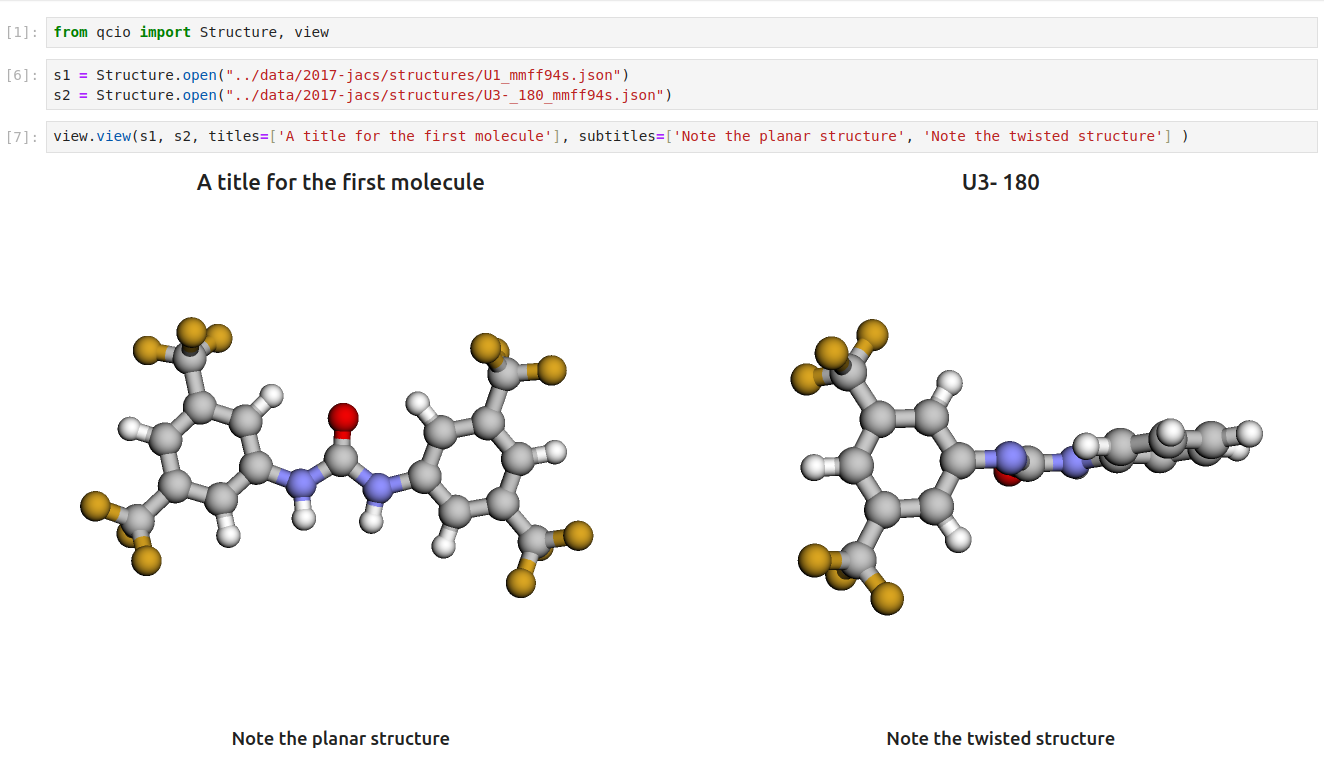
pip install 'qcio[view]'Then in a Jupyter notebook import the qcio view module and call view.view(...) passing it one or any number of qcio objects you want to visualizing including Structure objects or any ProgramOutput object. You may also pass an array of titles and/or subtitles to add additional information to the molecular structure display. If no titles are passed qcio with look for Structure identifiers such as a name or SMILES to label the Structure.
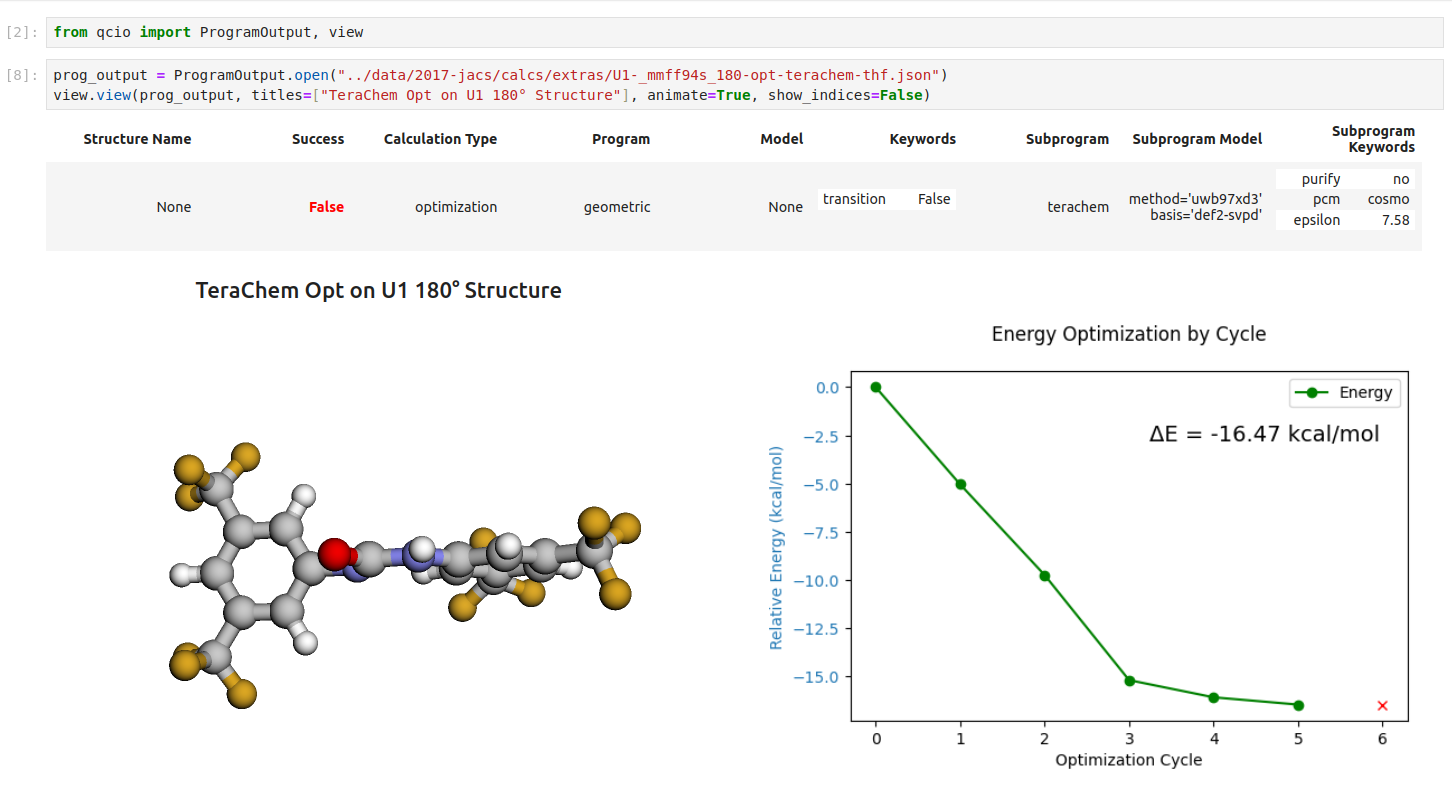
Seamless visualizations for ProgramOutput objects make results analysis easy!
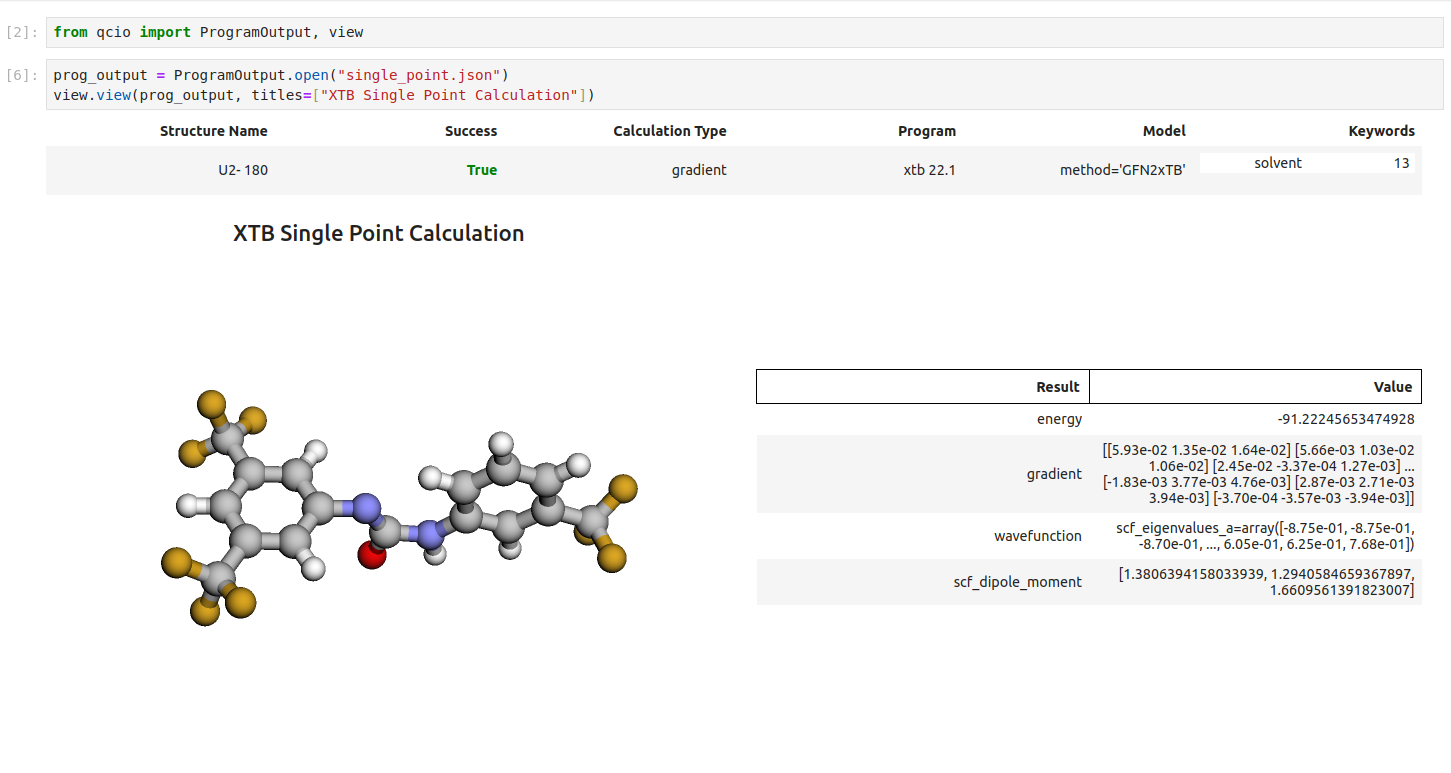
Single point calculations display their results in a table.
If you want to use the HTML generated by the viewer to build your own dashboards use the functions inside of qcio.view.py that begin with the word generate_ to create HTML you can insert into any dashboard.