A suite to handle single-cell electrophysiological data and to build and validate detailed electrical models.
- eFEL — Electrophys Feature Extraction Library
- BluePyEfe — Blue Brain Python E-feature extraction
- BluePyOpt — Blue Brain Python Optimisation Library
- BluePyMM — Blue Brain Python Cell Model Management
- BlueCelluLab - Blue Brain Cellular Laboratory
- EModelRunner — Runs cells from stand-alone packages
- BlueNaaS-SingleCell - Interacts with single cell models through a web application
- Currentscape - Plot currents in electrical models

Useful links: GitHub repo, Documentation.
Electrophys Feature Extraction Library
The Electrophys Feature Extraction Library (eFEL) allows neuroscientists to automatically extract features from time series data recorded from neurons (both in vitro and in silico). Examples are the action potential width and amplitude in voltage traces recorded during whole-cell patch clamp experiments. The user of the library provides a set of traces and selects the features to be calculated. The library will then extract the requested features and return the values to the user.

Useful links: GitHub repo, Documentation.
Blue Brain Python E-feature extraction
BluePyEfe aims at easing the process of reading experimental recordings and extracting batches of electrical features from these recordings. To do so, it combines trace reading functions and features extraction functions from the eFel library.

Useful links: GitHub repo, Documentation.
Blue Brain Python Optimisation Library
The Blue Brain Python Optimisation Library (BluePyOpt) is an extensible framework for data-driven model parameter optimisation that wraps and standardises several existing open-source tools.

Useful links: GitHub repo, Documentation.
Blue Brain Python Cell Model Management
When building a network simulation, biophysically detailed electrical models (e-models) need to be tested for every morphology that is possibly used in the circuit.

Useful links: GitHub repo, Documentation.
Blue Brain Cellular Laboratory
BlueCelluLab is designed for simulations and experiments on individual cells or groups of cells. Suitable use cases for BlueCelluLab include:
- Scripting and statistical analysis for single cells or cell pairs.
- Lightweight, detailed reporting on specific state variables after simulation.
- Developing synaptic plasticity rules.
- Validating dynamics of synaptic properties.
- Automating in-silico whole-cell patching experiments.
- Debugging, both scientifically and computationally.

Useful links: GitHub repo, Documentation.
Runs cells from stand-alone packages
EModelRunner is a python library designed to run the cell models provided by the Blue Brain portals in a simple and straightforward way.

Useful links: GitHub repo, Documentation
Interacts with single cell models through a web application
Blue-Neuroscience-as-a-Service-SingleCell is an open source web application. It enables users to quickly visualize single cell model morphologies in 3D or as a dendrogram. Using a simple web user interface, single cell simulations can be easily configured and launched, producing voltage traces from selected compartments.
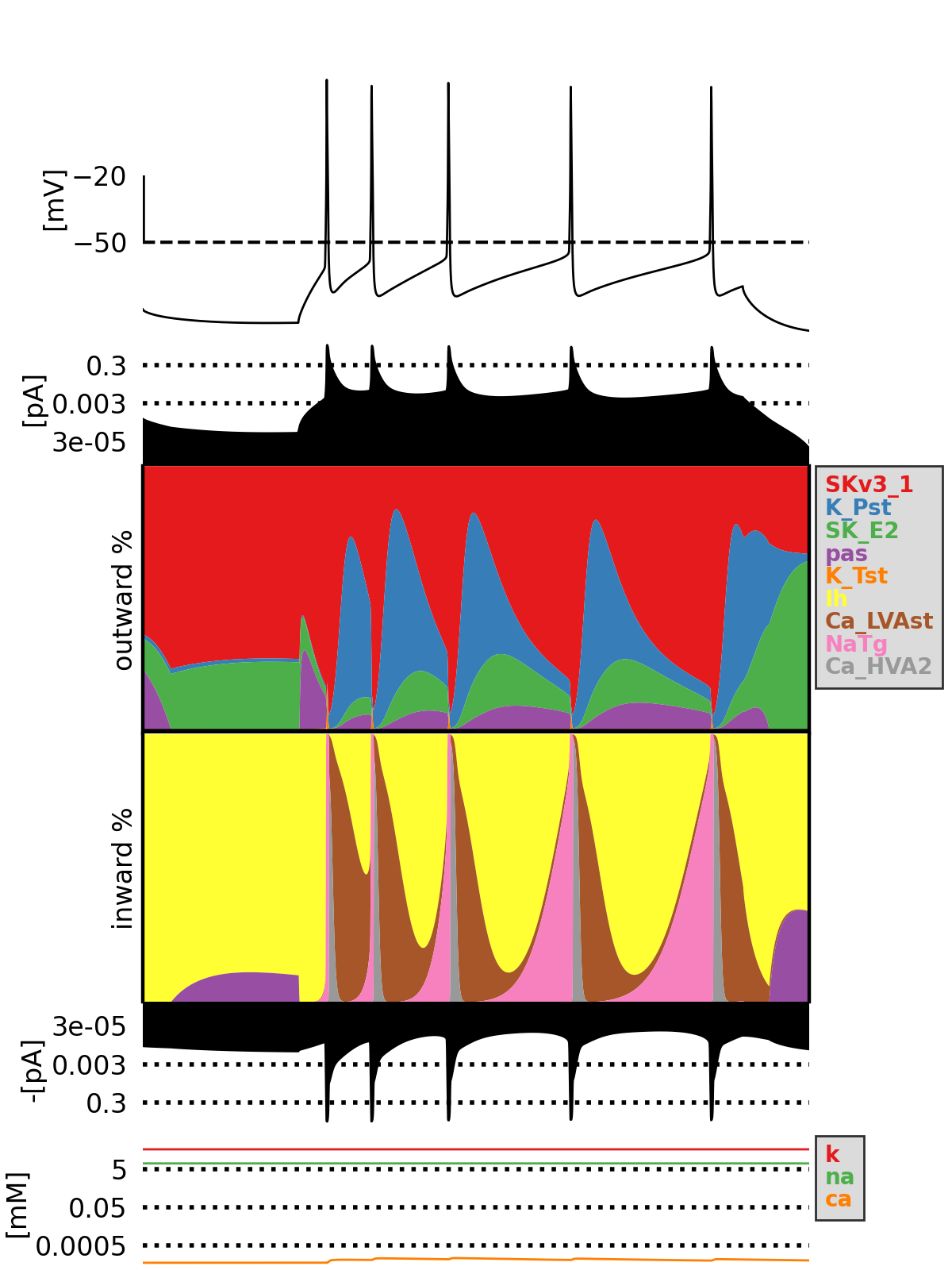
Currentscape is a Python tool enabling scientists to easily plot the currents in electrical neuron models. The code is based on the paper Alonso and Marder, 2019.
Useful links: GitHub repo, Documentation.