-
Notifications
You must be signed in to change notification settings - Fork 98
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
2 small South African clusters of Omicron/Delta recombinants with interesting Spike mutations (8 sequences) #844
Comments
|
Thanks @JosieLikesCats for flagging these at such an early stage, they are indeed very interesting sequences. I'm sorry, I can't really provide much insights yet, but I would very much appreciate if you could try to answer some questions that come to mind: Where in Limpopo and Gauteng are these from? Google maps says ~400km but could the Limpopo ones also be from a place closer to Gauteng province? These sequences should be run through sc2rf to check for recombination - first impression is that it's very recombinanty. But also that both sequences seem to have some things in common. Very speculatively, these could represent different results of intra host recombination - but one needs to look at this in more detail. It may be worth splitting this issue up and separate the proposals but for now to keep things simple I think it's fine to keep them both together. Here are screenshots from Nextclade to save everyone some time: |
|
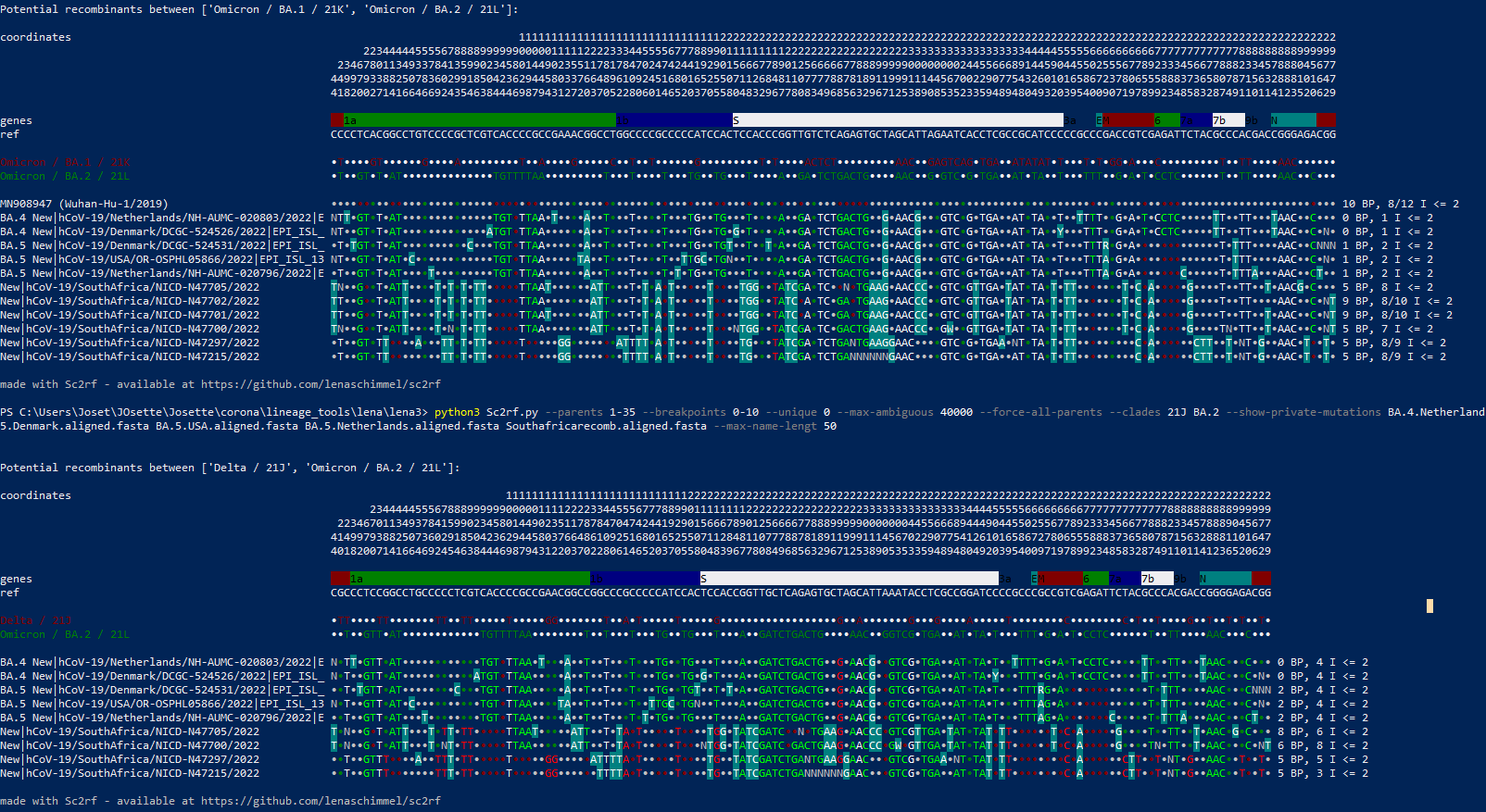
Just spotted these too, went to check and found this issue. These look like some weird combination of three different lineages: AY.45, a divergent BA.4/5, and BA.2 (seen near the 3' end where BA.4/5 mutations are absent from the Omicron-derived sections). Both have lots of apparent breakpoints but the 3' end of constellation 2 is particularly messy and seems to switch back and forth almost every mutation - the N protein alone goes Omicron/Delta/Omicron/Delta. If these are real I wouldn't be surprised if they are new emergences from the Omicron source, especially given the location. |
|
The small cluster (no 2) very much looks like a 21J and BA.4/5 recombinant to me - but with an unusually high number of 5 breakpoints. The ranges are as follows: |
|
Here is a spreadsheet comparing the mutations to AY.45, BA.2, BA.4 and BA.5: pango844.xlsx Constellation 1 has at least 6 breakpoints; constellation 2 has at least 5 as @corneliusroemer says. Could potentially be even more, as there are a couple of places where a single mutation or reversion could have been gained convergently or through recombination. I'm also unconvinced that BA.4/5 are involved - I think the Omicron parent may just be a divergent BA.2. Looking at all of the locations where BA.2 and BA.5 differ, referring to the constellations as C1 and C2 for short: If BA.5 is involved and not BA.2, we have to believe that the silent mutations at 12160 and 27889 both reverted, as well as S:F486V in C2. In my humble opinion, this seems much less likely than S:L452R and S:R493Q arising independently on top of BA.2 - after all, we've seen that in BA.2.77 too. The evidence against BA.4 is even stronger, e.g. neither constellation has the deletion in nsp1. |
|
Thanks @JosetteSchoenma -- here is a link to the UShER view with a permanently saved .json file that won't be deleted in a couple days, and with branches labeled by reversions/back-mutations: Since this is apparently a recombinant, UShER is not as useful as it might be otherwise. The phylogenetic tree assumes a steady accumulation of mutations, but recombinants violate that assumption. UShER places a recombinant sequence on the branch of the tree where it has the fewest differences from existing sequences, which usually corresponds to one of the parent lineages -- but there are reversions/back-mutations for the portions of the genome contributed by the other parent. (In fact, a long branch with multiple reversions is the signal that our RIPPLES tool uses to look for potential recombinants in the big tree... we should run that again one of these days!) These sequences are placed on a branch of BA.5 that is already riddled with various reversions that are probably mostly sequencing artifacts -- but the long branch makes it pretty clear that these sequences are different from the others, and that placement is just the best that usher could do given the circumstances, not necessarily an indication that the sequences on that subtree are closely related. |
|
I agree with @silcn's analysis and reasoning. One small thought to add - Regarding the silent mutation C22916A at S:452. |
|
Could this be an XT (or similar) + BA.5 recombination ? |
|
Possible CvSp query to catch both constellation 1 and 2: Edit it catches 7/7 sequences on Covspectrum based on the great list by @silcn |
|
Thanks for all analysis so far, very interesting to read all the comments! The final sequence has been uploaded to GISAID, so it should be available soon - I'll edit this comment once it's released. EDIT: sequence now released, EPI_ISL_13843609 @corneliusroemer to answer some of your questions: The sequences are from Johannesburg (Gauteng) and Polokwane (Limpopo), which are two of the main cities in each province. We have also had school and university holidays recently, and so there has likely been increased travel between provinces. I see recombination is being looked at quite closely by everyone, so I'll just add that we have some NGS-SA team members also taking a look with a variety of tools; we'll update accordingly if we find anything interesting. Thanks for adding the screenshots! I had considered two separate issues but thought for now since there are so few it made sense to keep it together. Happy to split these in future if needed. |
|
NGS-SA report on these sequences: |
|
I can confirm this query catches 7 out of 7 sequences of this new variant: |
|
There's a new sequence from Gauteng uploaded today clearly related to the others, though it's 29.7% NNN's—EPI_ISL_13913050 It has S:F486P and S:P621S, so it's part of constellation 1, but spike residues 1-340 and 670-1044 are blank according to NextClade. Unlike the other four sequences from constellation 1, this one has S:T572I. Collection date 2022-7-4. |
|
@ryhisner thanks for adding the EPI_ISL ID there, was just about to ask, and sorry the UShER web interface is using an old fasta-reading library that truncates names at the space character... "hCoV-19/South" is a pretty useless label. |
|
EPI_ISL_13913050 (SouthAfrica/CERI-KRISP-K045132/2022) is the first of these from outside NICD, and it is more similar to C1 than it looks in that UShER tree view -- it has Ns at 4456, 5869, 10198, 12163, 21623, and 23679, and it has C10954T and C28531T like NICD-N47701 and NICD-N47705, so I would expect UShER to place it at the end of that branch, instead of splitting it in the middle. Looking into why it didn't. Meanwhile here's a sc2rf view so you can see how CERI-KRISP-K045132 looks pretty much like the NICD C1 sequences, but with more Ns, and without reversions at 22686, 22688 and 22786 (common casualties of amplicon dropout I think): |
|
Although these clusters don't fulfill the minimum number of sequences - I think the extremely unique potential pattern of recombination might mean if sequences keep appearing they should get assigned as there will be justification for being able to refer to them by a non-ambiguous designation. Going to put a monitoring tag on this for now (hope thats okay @chrisruis @InfrPopGen !). |
|
Agree that it's worth seeing if any new sequences in this cluster appear and if they do to designate. The minimum number of sequences is not a hard limit, we can make exceptions if there are good reasons (there are here). |
|
Hi everyone, just a heads-up that one more sequence from constellation 2 will be released in the next couple of hours (N46078, EPI_ISL_14112354). Also from Limpopo, with collection date of 30 May. We haven't yet detected any more recent samples but are monitoring closely. |
|
The new constellation 2 sequence is missing M:R146H and so will not be picked up by @FedeGueli's cov-spectrum query. Here is a query that will pick up everything once cov-spectrum is updated with the new sequence: |
|
thx @silcn checked your new query 9 out of 9! |
|
2 more sequences from Constellation 1: EPI_ISL_14585888, EPI_ISL_14585891 |
|
Great! I wouldn't make these sublineages of each other as there may be
recombination involved. I'd just call this XAV, and the next one XAW or
whatever :)
…On Fri, Aug 26, 2022, 13:16 Angela Sun ***@***.***> wrote:
This seems like the appropriate thing to do - I'll reopen the issue and
monitor for further development in constellation 2, and designate the
larger constellation as XAY.1.
—
Reply to this email directly, view it on GitHub
<#844 (comment)>,
or unsubscribe
<https://github.com/notifications/unsubscribe-auth/AF77AQIHR6CUTU7Y4OEUUVLV3CRQLANCNFSM53TGFWRA>
.
You are receiving this because you were mentioned.Message ID:
***@***.***>
|
|
I'll leave the first cluster as XAY then! |
|
One more sequence i think popped up: EPI_ISL_14728611 Gauteng i think it is XAY (Constellation 1 ) :S:A706V,F486P, P621S, R21G, |
|
Added this to XAY. |
|
Gisaid query M_R146H, Spike_F186L for XAY |
@FedeGueli I think that query covers both C1 (designated XAY) and C2 (monitored). Extending your query for XAY: for C2: |
|
Thx @AngieHinrichs !! yes i usually do the S:F486P and then the other! good to have them separately thank you very much |
Added new recombinant lineage XBA from #844 with 4 new sequence designations, and 0 updated designations
|
Lineage XBA has been designated for constellation 2, with four example sequences. The lineage alias is given as an interim AY.45/BA.2 recombinant, with one breakpoint, because that at least gives pipelines what they expect when reading the json. |
|
New XAY just uploaded an elderly man in Cape Town collected on 31/08/22. EPI_ISL_14975893 |
|
The first international XAY has just appeared in Denmark:
Travel information is not available, but we know this is a reinfection with last infection in January, possibly BA.2 given this was Denmark. |
|
There is another XAY from Denmark. Getting the same place on the Usher tree as the one @corneliusroemer mentioned. 2nd of October 2022. EPI_ISL_15284246. |
|
Two XAY more sequences from south africa have been uploaded collected on 29/8 and 14/9 both from Gauteng and baseline surveillance. |
|
i think i have found a better query for XAY : Spike_P621S,Spike_F186L , it actually founds 27 sequences from 3 countries while Gisaid Pangolin calls 23 viruses XAY and our old manual query a lot less. |
|
Thanks @FedeGueli, that helped me find a couple new CA sequences that were being excluded from the tree but should be added tomorrow! Also missing but hopefully added tomorrow: SouthAfrica/SU-NHLS_5859/2022|EPI_ISL_14975893|2022-08-31 |
|
Found a sequence that Nextclade sees as XBA but Usher puts outside every branch starting directly from the B.1.1.529 root, bu it is mixed between Delta and Omicron. It is from Belgium and sampled recently: EPI_ISL_15537619 @corneliusroemer @thomasppeacock @AngieHinrichs @JosieLikesCats @JosetteSchoenma @c19850727 @silcn @shay671 |
|
Command-line nextclade places it with XBA as the closest match... but with 21 reversions relative to the XBA placement, as well as 7 mutations associated with other clades, and 28 additional mutations. It's excluded from the UShER tree because it's Omicron-ish but so divergent from its nextclade placement. My guess is contamination, but that's just my guess based on looking at nextclade numbers; someone looking at the raw data might see something else. |
|
It looks like another recombinant strain that is related to XAY/XBA to me. The reversions can be easily explained by different breakpoints. This one is an XBA-like with a Delta-like S2. The S2 part (P681R+V736I+T859N+D950N) looks pretty real. T859N is one of the most notable convergent mutations in the late Delta era. |
|
This one also may give some hints about how XAY/XBA evolved. This one has L452M, elaborating the L452M->R theory. |
|
No, this one is unrelated to XAY/XBA. Orf1b:M115I and C25413T from AY.45 is missing. It is another Omicron/Delta recombinant that is strikingly similar to XBA. |
|
A breaking point between S:EFR156G and S:V213G like XAY/XBA, which is also close to XAW and XBC breaking points. |








Hi everyone, I'm just opening this issue to highlight that there are several sequences with an unusual mutation pattern in our most recent upload from South Africa, which will potentially represent two new lineages if more sequences are detected. The teams in our genomic surveillance network (NGS-SA) as well as our public health institute (NICD) are closely monitoring the sequences and cases in the country. These new constellations have been detected only in a small proportion of recent data, and our cases remain low.
I know these do not yet meet requirements for designation, as there are only N=4 and N=3 (2 available on GISAID, last 1 will be released tomorrow) sequences for each constellation, but we thought they would probably be of interest and picked up here/on Twitter eventually. For now, please see below for some details and the major mutation profiles for the two groups of sequences.
N=4 constellation 1
Earliest sequence: 28 June 2022
Most recent sequence: 29 June 2022
Circulating: Gauteng, South Africa
Nextclade assigns 21M but flags lots of private mutations (mainly 21J), pango assigns Unassigned/B.1.1.529
Genomes
EPI_ISL_13830378
EPI_ISL_13830377
EPI_ISL_13830376
EPI_ISL_13830375
N=3 constellation 2
Earliest sequence: 13 June 2022
Most recent sequence: 24 June 2022
Circulating: Limpopo, South Africa
Nextclade assigns 21J but flags lots of private mutations (mainly 21K/21L), pango assigns XD
Genomes
EPI_ISL_13830379
EPI_ISL_13830380
Evidence
constellation1_defining_aa_changes.xlsx
constellation2_defining_aa_changes.xlsx
Spike mutations in constellation 1 only, relative to Omicron: R21G, F486P, P621S, A706V
Spike mutations in constellation 2 only, relative to Omicron: S477D
Shared mutations relative to Omicron BA.4/5: L18F, T19R, W152L, E156del, F157del, R158G, F186L, G446D, T1117I
Notably both clusters have a second silent nt change in L452R not present in BA.4/5.
There are some significant differences outside spike (see attached mutation profiles).
The sites 213, 371, 373, 375, 376, 408, and 764 are not reliably covered by the data, so they cannot be confirmed yet.
UShER tree (including 7th sequence to be uploaded): https://nextstrain.org/fetch/genome.ucsc.edu/trash/ct/singleSubtreeAuspice_genome_1ef08_5f410.json (in a previous Usher tree they clustered near XD).
The text was updated successfully, but these errors were encountered: