Create, edit and use hierarchical taxonomies in Plone.
This add-on provides support for hierarchical taxonomies in multiple languages, letting users easily associate existing content with terms from one or more taxonomies.
Taxonomy is the "discipline of defining groups [...] on the basis of shared characteristics and giving names to those groups. Each group is given a rank and groups of a given rank can be aggregated to form a super group of higher rank and thus create a hierarchical classification".
Here's an example of the "taxonomic kingdoms of life":
Living Organisms Living Organisms -> Eukaryotic Living Organisms -> Eukaryotic -> Simple multicells or unicells Living Organisms -> Eukaryotic -> Multicellular Living Organisms -> Eukaryotic -> Multicellular -> Autotrophic Living Organisms -> Eukaryotic -> Multicellular -> ... Living Organisms -> Prokaryotic Living Organisms -> Prokaryotic -> Archaebacteria Living Organisms -> Prokaryotic -> Eubacteria Living Organisms -> Prokaryotic -> Eubacteria -> ...
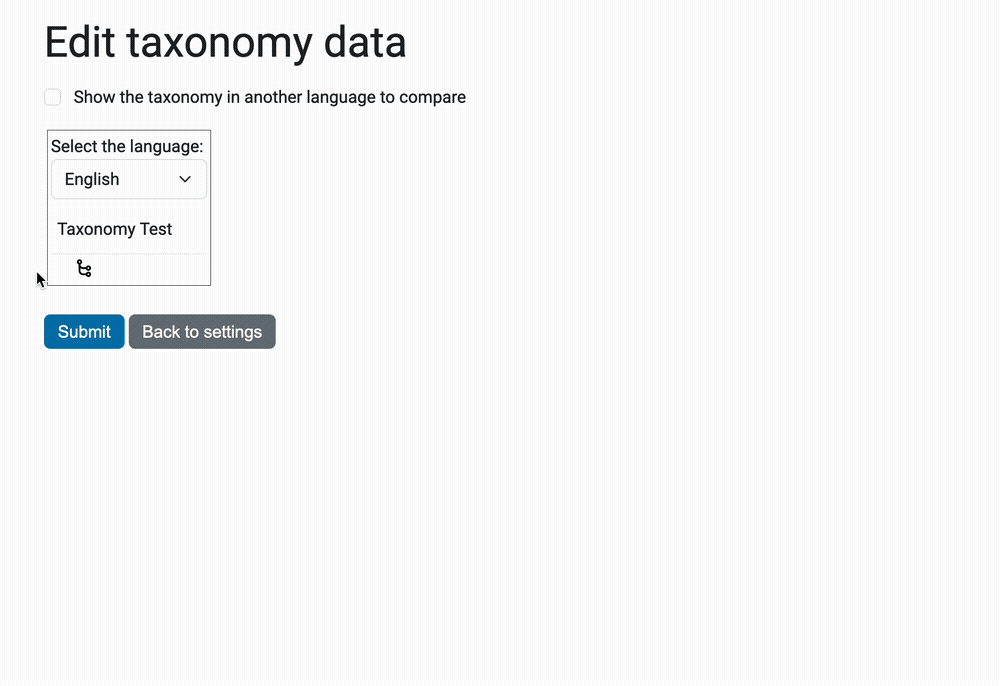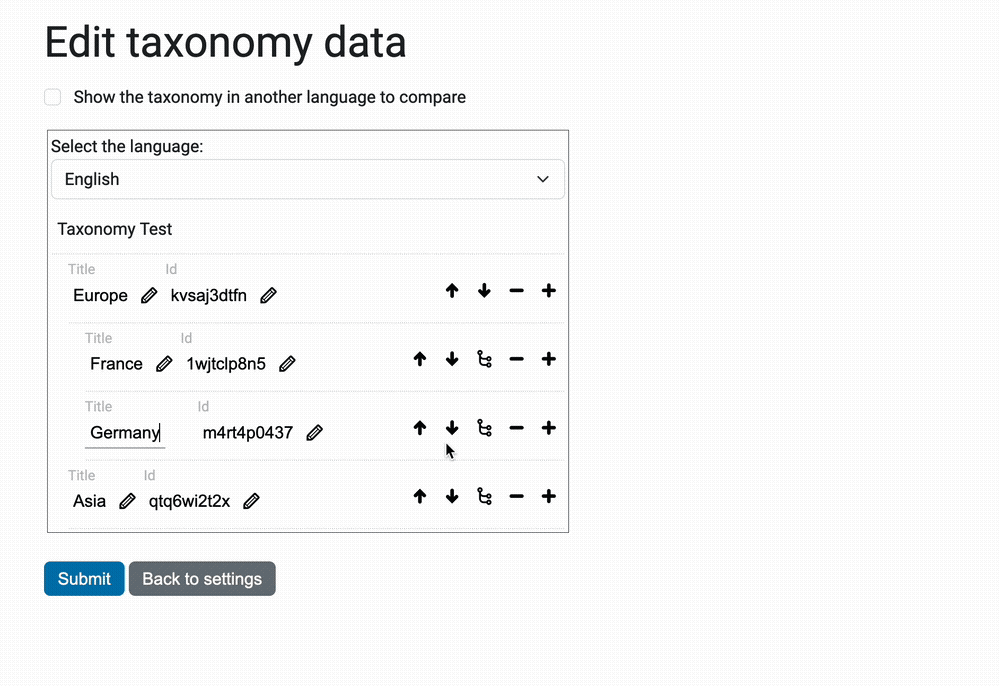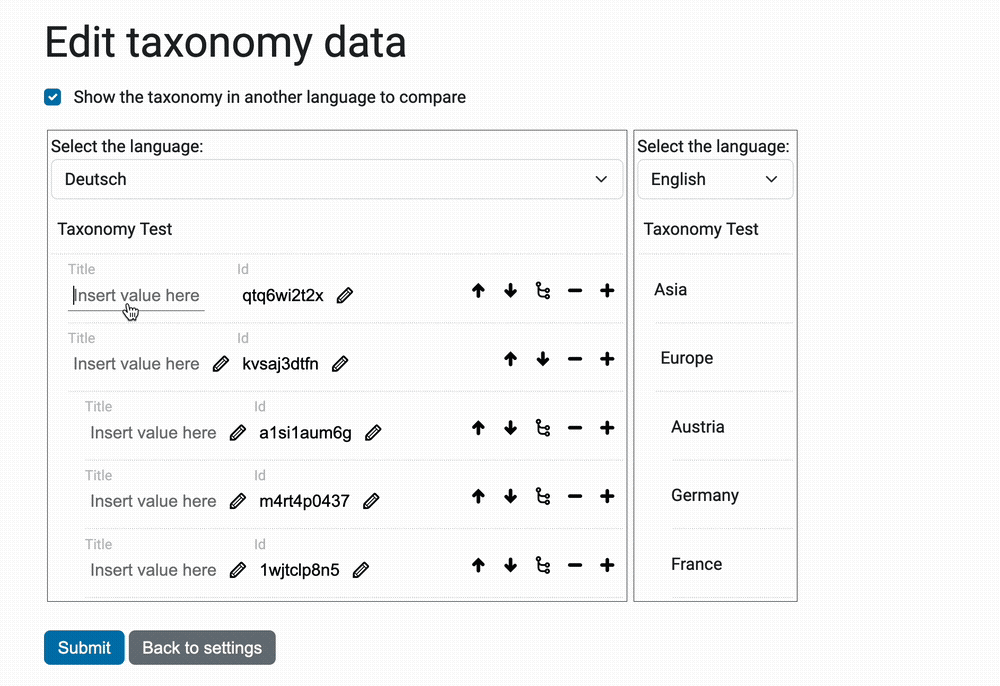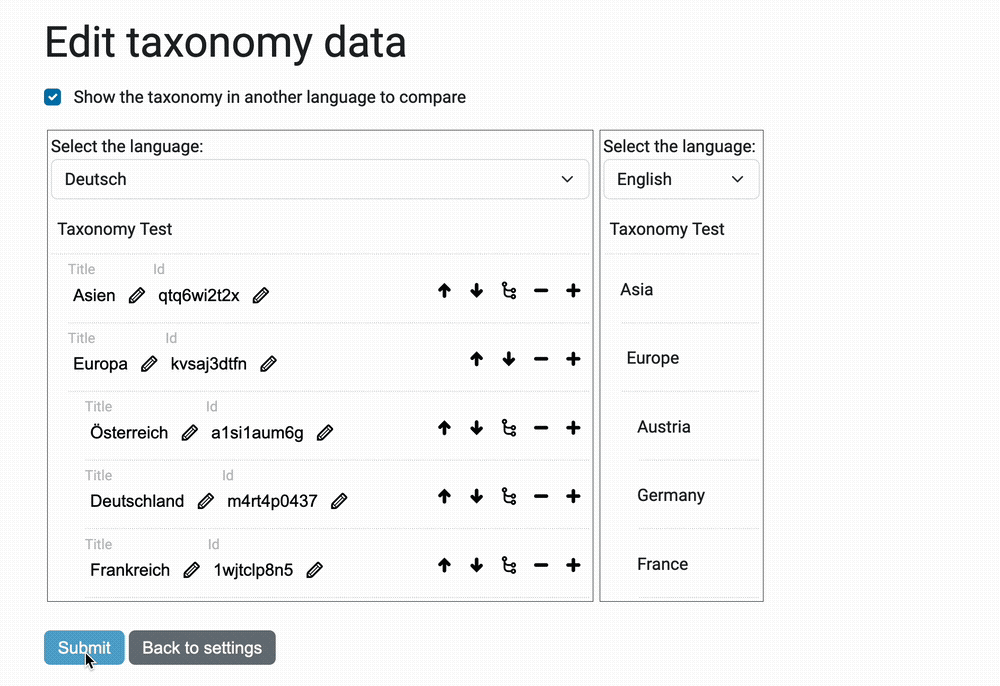
Taxonomies can be quite large, sometimes in the tens of thousands (10,000+). And in sites with multiple languages, each title – or caption – must appear in translation.
Note that the selection of a term in the hierarchy implies the selection of all its parents. In the example above this means that if "Eubacteria" is selected, then also "Prokaryotic" and "Living Organisms" will be.
The implementation tries to meet the following requirements:
- Support many (10,000+) terms.
- Terms can be organized in a hierarchical classification.
- Easily import and export in a common format (VDEX).
- Taxonomies will provide vocabularies that are translatable.
- Use behaviors to provide a choice field for each taxonomy.
- Manage taxonomies and assign to content types "through-the-web".
In the description below, we touch on each of these requirements.
In order to limit both the memory and computation requirements, the term data is contained in exactly one persistent index per language, a mapping from the materialized term path to its term identifier.
The term:
Living Organisms -> Eukaryotic -> Simple multicells or unicells
will be indexed under this path:
"Living Organisms/Eukaryotic/Simple multicells or unicells"
The index allows us to provide an iterator over the sorted vocabulary terms, virtually without cost (as well as containment queries).
At the same time, while the hierarchy is encoded, we can quickly look up terms in a subtree.
Note: As collective.taxonomy uses slash as separator, you have to
monkey patch the PATH_SEPARATOR constant if you want to use '/' in
your terms.
While collective.taxonomy (this package) does make it possible to
create, manage and edit taxonomies from a browser-based interface, the
primary focus is to support the exchange of terms in the VDEX format:
The IMS Vocabulary Definition Exchange (VDEX) specification defines a grammar for the exchange of value lists of various classes: collections often denoted "vocabulary".
This exchange is integrated with GenericSetup which manages imports and exports using setup profiles. It is also possible to use the control panel for importing and exporting VDEX files.
The package comes with integration for the Dexterity content type framework:
for each taxonomy, a behavior is available that adds a choice field
which pulls its vocabulary from the taxonomy. The behavior is
configurable in terms of field name, title and whether it allows the
selection of one or more multiple terms. You should first install
dexterity and then collective.taxonomy, otherwise the behaviors
for the existing taxonomies will be missing.
The main objective during this project has been to get a high rate
of through-the-Web administration. Therefore the use of the product
will not require any Python programming nor configure.zcml directives.
In the control panel (/@@taxonomy-settings), the user can:
- Import taxonomies from VDEX files.
- Export taxonomies existing to VDEX files.
- Delete taxonomies
- Add and delete behaviors for taxonomies
When a new behavior is created for a taxonomy, it can easily be added
to the desired content types using the content type control panel, provided
by Dexterity. After this is done, the taxonomy is available on add and edit
forms, and it is also available for collections, if plone.app.collection
is used on the site. An index is also created, so the taxonomies can easily
be used for catalog queries.
Please read the detailed Getting Started Tutorial
The view @@taxonomy-edit-data that allow users to edit the taxonomy data
is a React/Redux app (the source code is in the javascripts directory.
Here's a preview of this view:
The languages allowed for the taxonomies are the languages defined in portal_languages.
All JS development resources are in the directory resources/js.
While making changes to these files you can:
- change to directory
resources/- run
make serve
This starts a webpack development server at localhost:3000 and
serves the JS files.
Now start your Plone instance with
NODE_ENV=development bin/instance fg
and the taxonomy controlpanel will use the files served from the webpack server.
When you have finished your changes run:
make test make build
this will build the resources and save it in src/collective/taxonomy/static/js.
The app uses react-intl to handle i18n.
To translate the app, add a new language in the translations directory. For example,
create a es file in the translations directory that contains:
const es = {
submitLabel: 'Enviar',
}
export default es
Then, edit translations/index.js to add the language to the translations object:
import es from './es'
const translations = {
es,
fr
}
You'll have to rebuild the js bundle: npm run build
That's it!
This package also supports endpoints for basic CRUD operations that can be consumend by any frontend service.
The API consumer can create, read, and delete taxonomies.
| Verb | URL | Action |
|---|---|---|
| POST | /@taxonomy | Add taxonomy with specific data |
| GET | /@taxonomy | List all taxonomies |
| GET | /@taxonomy/{name} | Get taxonomy data |
| DELETE | /@taxonomy/{name} | Remove one or more taxonomies |
| PATCH | /@taxonomy/{name} | Update taxonomy data |
| GET | /@taxonomySchema | Get current taxonomy schema |
This product has been translated into
- Danish.
- German.
- French.
- Spanish.
You can contribute for any message missing or other new languages, join us at Plone Collective Team into Transifex.net service with all world Plone translators community.
collective.taxonomy version 3.x
- Plone 6.1 (py3)
- Plone 6.0 (py3)
collective.taxonomy version 2.x
- Plone 5.2 (py2/py3)
- Plone 5.1
- Plone 5.0
collective.taxonomy version 1.x
- Plone 4.3
- or an older version using a recent version of plone.dexterity/plone.app.dexterity
How can I import an existing ATVocabularyManager vocabulary?
Use the script provided in this gist. Just remember to edit the vocabIdentifier and vocabName.
Have an idea? Found a bug? Let us know by opening a ticket.
- Issue Tracker: https://github.com/collective/collective.taxonomy/issues
- Source Code: https://github.com/collective/collective.taxonomy
- Bo Simonsen <[email protected]>
- Malthe Borch <[email protected]>
- Thomas Clement Mogensen <[email protected]>
- Thomas Desvenain <[email protected]>
- Maurits van Rees <[email protected]>
- Cédric Messiant <[email protected]>
- Leonardo J. Caballero G. <[email protected]>
- Peter Mathis <[email protected]>
In 2010, Rok Garbas <[email protected]> reimplemented and extended prior work by Seantis and released collective.vdexvocabulary. This package allows you to configure and populate vocabulary components from a VDEX-specification. The package supports flat vocabularies only, and support for multiple languages takes a different approach (vocabularies are returned in an already translated form). Note that vocabularies are loaded in a read-only mode, although it's been proposed that vocabularies might be edited through-the-web.
In 2005, Jens Klein <[email protected]> released ATVocabularyManager. This package makes it possible to create taxonomies using Plone's content management interface with terms existing as regular site content. It's integrated with the Archetypes content type framework (now deprecated).
| [1] | Term relationships are currently not supported. |
The project is licensed under the GPL v2 or later (GPLv2+).