-
Notifications
You must be signed in to change notification settings - Fork 5
Differences to Ferguson Model
This page details the location and nature of any changes we have made to the Ferguson Model in our implementation, where it was not possible/desirable to reimplement the original version exactly.
Location: pyEpiabm/pyEpiabm/test_file.py
Implementation: We bulk generate random numbers at the start of the simulation, instead of generating them on the fly.
Motivation: On the fly random number generation is much slower in python than C++ so this had become prohibitively expensive.
Behaviour: This should not change the behaviour of the model.
Validation: This has not been explicitly validated against the Ferguson implementation.
Location: pyEpiabm/pyEpiabm/sweep/host_progression_sweep.py, the _set_infectiousness method.
Ferguson: Defines the infectiousness of a symptomatic infected person as negative.
Alternative: We differ and conserve the infectiousness as a positive number. The difference between the infectiousness of an asymptomatic or symptomatic infected person will be asserted in a different way.
Motivation: It seems more natural and clearer approach, thus a better implementation.
Behaviour: It should not affect the behaviour of the rest of the model, as the difference between the infectiousness of an asymptomatic vs symptomatic infected person will be measured using the infection status attribute of the person.
Location: pyEpiabm/pyEpiabm/sweep/host_progression_sweep.py, in the __init__
Ferguson: Scales the infectiousness progression array with its average, resulting in small differences in the scaling parameter with varying time steps length.
Alternative: We use the average of the input infectiousness profile to scale the infectiousness progression for each time step length.
Motivation: It reduces the variation in the infectiousness values for different time step lengths.
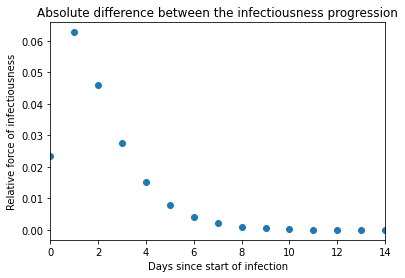
Behaviour: It has a minimal impact on most of the infectiousness values in the progression, except for the values in the peak of infectiousness where the new method overestimates a bit the infectiousness (a difference of 0.06 on the peak value when the length of the time steps is 1 day, where the CovidSim method estimates the infectiousness at 4.592 whereas our method estimates the infectiousness at 4.655). The absolute difference between infectiousness for a simulation of a time step length of 1 is illustrated below.
Location: pyEpiabm/pyEpiabm/travel_sweep.py, the sweeper() function for introducing infected individuals.
Ferguson: Implement a TravelDepartSweep and TravelReturnSweep using airport information.
Alternative: We introduce infected (mild or asymptomatic) individuals at every time step of the simulation based on a ratio of the infected individuals in the population as we assume that if many people are infected in the population the chance that an individual introduced to the population is infected is higher. Besides, constant number of individuals at every times step, or specific for each time step, can be introduced alongside or on its own. These travelling individuals are assigned to microcells with high population density. They are assigned to an existing household or will form their own household within this microcell. Travellers will leave the population after a specified duration of their stay (as long as they are not in isolation/quarantine).
Motivation: More general implementation of travellers that are infectious.
Behaviour: This allows for greater readability and flexibility within the model.
Location: pyEpiabm/pyEpiabm/intervention/ for the interventions, and pyEpiabm/pyEpiabm/sweep/intervention_sweep.py to generate the intervention sweep.
Ferguson: Interventions are active based on time and a specific trigger. This trigger is flexible defined and could either be the number of confirmed cases, number of critical cases that require intensive care or the number of deaths. For all interventions the type of trigger should be the same. Triggers can be set by admin unit (regional) or at a country (global) level.
Alternative: The only implemented trigger type here is the number of cases. This trigger can only be set on a global level.
Motivation: This is a basic first implementation of the interventions.
Location: pyEpiabm/pyEpiabm/intervention/case_isolation.py to carry out case isolation, and pyEpiabm/pyEpiabm/sweep/intervention_sweep.py to activate and deactivate the intervention.
Ferguson: Upon case detection case isolates with a certain probability. If this isolated person is a child that requires supervision an adult will stay home as well.
Alternative: Individuals will isolate with a certain probability when they are symptomatic or when they test positive. Adults staying home to take care of their child is not (yet) implemented.
Motivation: Isolating individuals based on symptoms allows for simulating scenarios in which tests are not available. Adult supervision of children could be a future implementation of the model.
Location: pyEpiabm/pyEpiabm/intervention/travel_isolation.py to carry out travel isolation, and pyEpiabm/pyEpiabm/sweep/intervention_sweep.py to activate and deactivate the intervention.
Ferguson: Travelling is captured in the TravelDepartSweep and TravelReturnSweep which take air travel and hotels into account.
Alternative: Infected individuals introduced to the population will isolate in an existing household or in a hotel when Travel Isolation is active. Isolating travellers can only leave the population after their isolation duration has passed.
Motivation: The specific Travel Isolation intervention allows for direct isolating infected travellers in a simplistic way.
Location: pyEpiabm/pyEpiabm/intervention/household_quarantine.py to carry out household quarantine, and pyEpiabm/pyEpiabm/sweep/intervention_sweep.py to activate and deactivate the intervention.
Ferguson: People who share household with a symptomatic case stay home. Infectiousness of the quarantined individual is scaled at household level. Infectiousness and susceptibility of the quarantined individual is scaled at place level and spatial level with their specific parameters.
Alternative: Household Quarantine is an additional measure on top of Case Isolation. People who share household with a case isolating individual stay home. The susceptibility of these susceptible individuals is scaled at household, place, and spatial levels.
Motivation: Implementing Household Quarantine on top of Case Isolation allows for specific treatments for the infected and susceptible individual within the household.
Location: pyEpiabm/pyEpiabm/intervention/place_closure.py to carry out place closures, and pyEpiabm/pyEpiabm/sweep/intervention_sweep.py to activate and deactivate place closures.
Ferguson: Generate an approach to close places based on their place types. Place closure subsequently scales the households' infectiousness, infectors' spatial infectiousness, potential infectees' spatial susceptibility and scales place infectiousness to 0.
Alternative: We follow the Ferguson model to build place-type dependent closures and scale down the susceptibility and infectiousness when the indicated place types are closed. We diverge from the Ferguson model in terms of ignoring the minimum radius for place closure and the proportion of places remaining open after closure by place type. The primary reason is that actual interventions mostly close all place types simultaneously, for example, closing all schools rather than only part of them. Besides, rather than providing a list to change levels of place closure, we allow closure events with different scaling parameters.
Motivation: This change better mocks the realistic intervention and simplifies the model by removing unnecessary parameters. The model is more reasonable as it requires a full list of parameters rather than fixing the scaling parameters for different events.
Location: pyEpiabm/pyEpiabm/intervention/social_distancing.py to carry out social distancing, and pyEpiabm/pyEpiabm/sweep/intervention_sweep.py to activate and deactivate social distancing.
Ferguson: Generate an approach to distance individuals in society. They may perform enhanced social distancing based on their compliance, which is associated with their age groups. Social distancing subsequently scales the susceptibility of individuals, households, and places.
Alternative: We follow the Ferguson model to build age-dependent social distancing and scale down the susceptibility when this intervention starts. We diverge from the Ferguson model in terms of ignoring the minimum radius for social distancing. We also remove the overall proportion of compliance with enhanced social distancing as the same effects can be reached by specifying it in each age group. Similar to place closure, we allow social distancing with various scaling parameters at different times.
Motivation: This change better simplifies the model by removing unnecessary parameters and allows more complex situations by providing a complete list of parameters rather than fixing the scaling parameters for different events.
Location: pyEpiabm/pyEpiabm/intervention/disease_testing.py to carry out testing, and pyEpiabm/pyEpiabm/sweep/host_progression_sweep.py to generate the testing queue.
Ferguson: Testing is implemented in the context of digital contact tracing, however there is no general implementation of disease testing.
Alternative: We include an explicit disease testing framework that allows both symptomatic and asymptomatic individuals, as well as those who are uninfected, to be tested. The proportion of individuals who go for tests can be controlled according to whether they are symptomatic or not, varied according to whether the person is a care home resident, key worker, or other, and separated into PCR or LFT testing flows in given proportions. On testing, individuals may be a false positive or false negative, with probabilities specified for PCR and LFT testing separately.
Motivation: A general implementation of disease testing.
Behaviour: Testing can be connected to case isolation such that positive individuals isolate. In addition this framework allows the effect of false positive and false negatives on infection curves and societal impact to be evaluated.
Location: pyEpiabm/pyEpiabm/intervention/vaccination.py to carry out vaccinations, and pyEpiabm/pyEpiabm/sweep/initial_vaccine_sweep.py to generate the initial vaccination queue.
Ferguson: Generate an age-based priority queue for vaccination. Vaccination subsequently scales a persons infectiousness and susceptibility.
Alternative: We follow the Ferguson model in allowing for age-prioritised vaccination, but also prioritise care home residents. We also similarly define a vaccination capacity and vaccinate a number of individuals per day according to that capacity. The infectiousness of a vaccinated individual is reduced. We diverge from the Ferguson model in terms of how vaccination affects susceptibility. A proportion of vaccinated individuals develop complete immunity, while others remain susceptible to infection.
Motivation: This change better represents the protection offered by vaccination and allows us to use more readily available parameters.
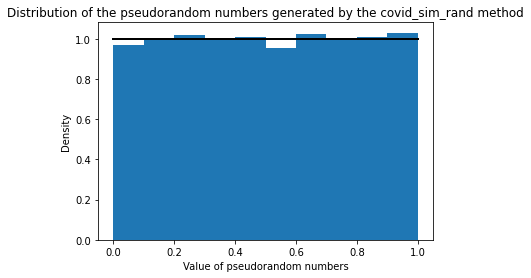
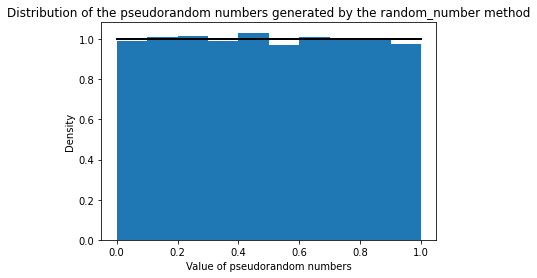
Location: pyEpiabm/pyEpiabm/utility/random_methods.py and pyEpiabm/pyEpiabm/sweep/host_progression.py
Change: We use random.random() to generate pseudorandom numbers instead of using the implemented covid_sim_rand(), the equivalent of what is used in Ferguson's model.
Motivation: It is simpler and clearer.
Behaviour: This should not change the behaviour of the model.
Validation: We compared the distribution of the generated pseudorandom numbers from the covid_sim_rand() method with the ones generated by random.random() function. The results convincingly show that the two methods are equivalent.
Location: pyEpiabm/pyEpiabm/utility/inverse_cdf
Change: We implemented two similar choose method that can be found in CovidSim, one without exponentialisation and one with negative exponentialisation on which is applied the natural logarithm and then multiplied by one. For simplicity, in the program we use the method without exponentialisation.
Motivation: It is simpler and should not change the result.
Behaviour: It does not change the resulting sampling.
Validation: We compared below the distribution of sampling for latent time for the two methods. The results are very similar and do not seem to be significantly different.


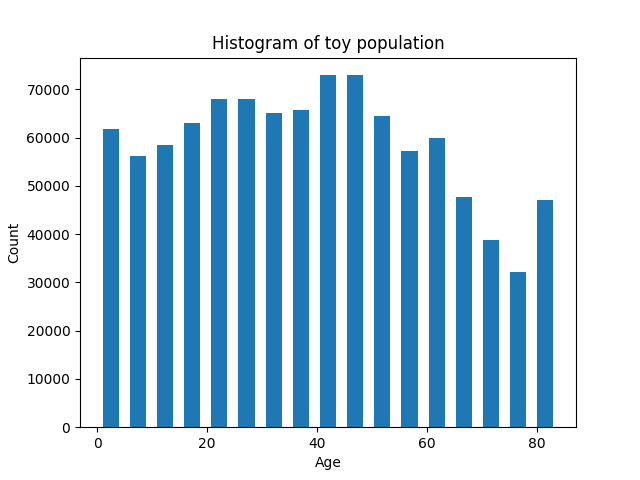
An error was found in the initial_household_sweep.py file which changed the chance of people being added to households by using incorrect conditional probability. A random number between 0 and 1 was generated and compared against different probabilities. The first comparison was correct (r < a) but when a second case was considered the previous code checked r < b/(1-a). This leads to a too small chance of the second condition being satisfied as we have already checked if r < a so the condition we are checking for is a < r < b/(1-a) rather than a < r < b. To see this clearly let's use some dummy values, a = 0.1 and b = 0.3. Then if r<0.1 this is correct however if r > 0.1 then the CovidSim condition is r < 0.3/(1-0.1) which is r < 1/3. However we know that r > 0.1 so the condition is only satisfied if 0.1 < r < 1/3. This error was carried forward from CovidSim so this is a deviation from CovidSim. We have changed the equations for the if statements to rectify this situation. We have also written histogram plotting code in the age_stratified_example to show the effect.
This population is still not correct but this is due to a combination of parameter values for assigning people to households. The greatest error is in 5-15 year olds (there are too many of them) and a few too few 20-30s. This is probably due to the incorrect assumption that for any household of 4 or more there MUST be children. This leads to too many children being added as well as a few too few young adults being added as they cannot be parents of children of this age in our parameter assumptions (19 year age difference needed).
The new histograms we get for the population are below.

Location: `pyEpiabm/pyEpiabm/sweeps/spatial_sweep.py
Change: We weight the choice of people to be selected by the model to infect by the inverse of the distance between 2 cells multiplied the number of people in the given cell. This means that individuals in less dense cells are not over-represented in the selection of individuals to infect spatially. We also in this way skip over cells with 0 people in them
Motivation: It is more accurate.
Behaviour: This should improve the spatial behaviour of the model.
Validation: Simulations affect closer cells more strongly than cells that are further from the infection point. Cells with 0 people no longer break the code