- Data and methods to replicate the results obtained by Simpati, netDx and PASNet
- Install Docker for your operating system following the link
- Pull two containers (one containing Simpati enviroment, one netDx and one PASNet): lgiudice/simpati:latest and lgiudice/comp1:latest and lgiudice/comp2:latest
- With bash:
docker pull lgiudice/simpati:latest docker pull lgiudice/comp1:latest docker pull lgiudice/comp2:latest
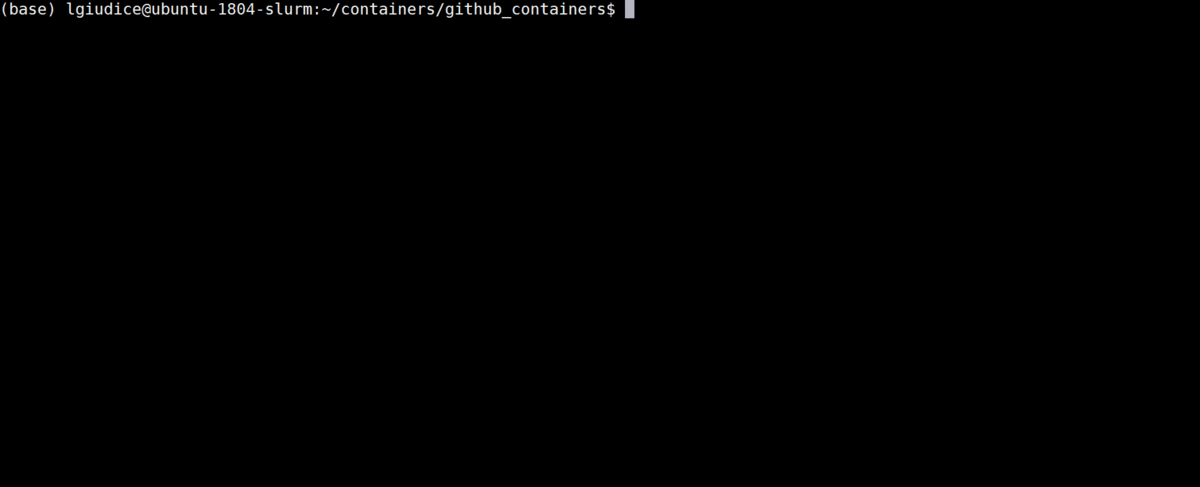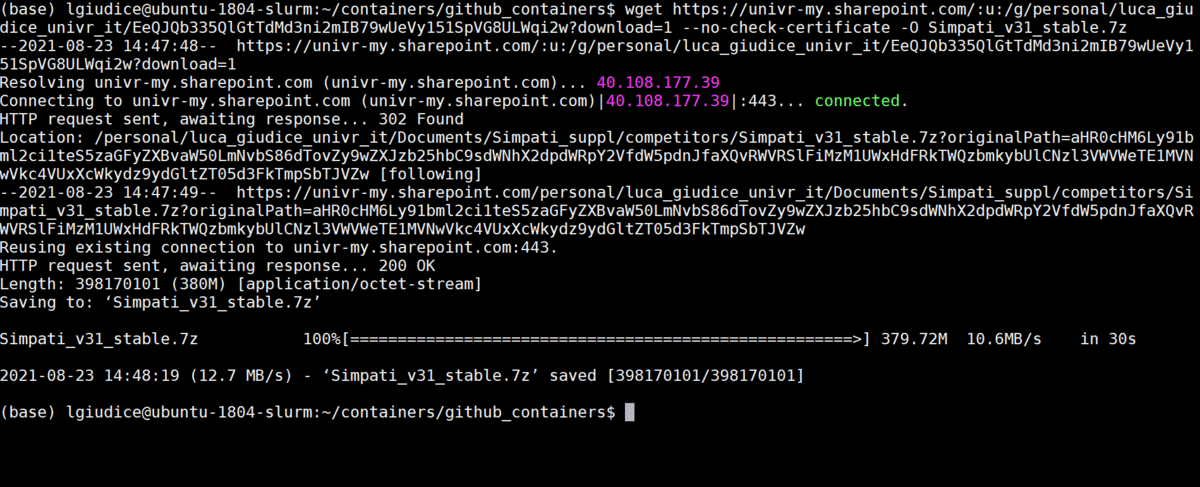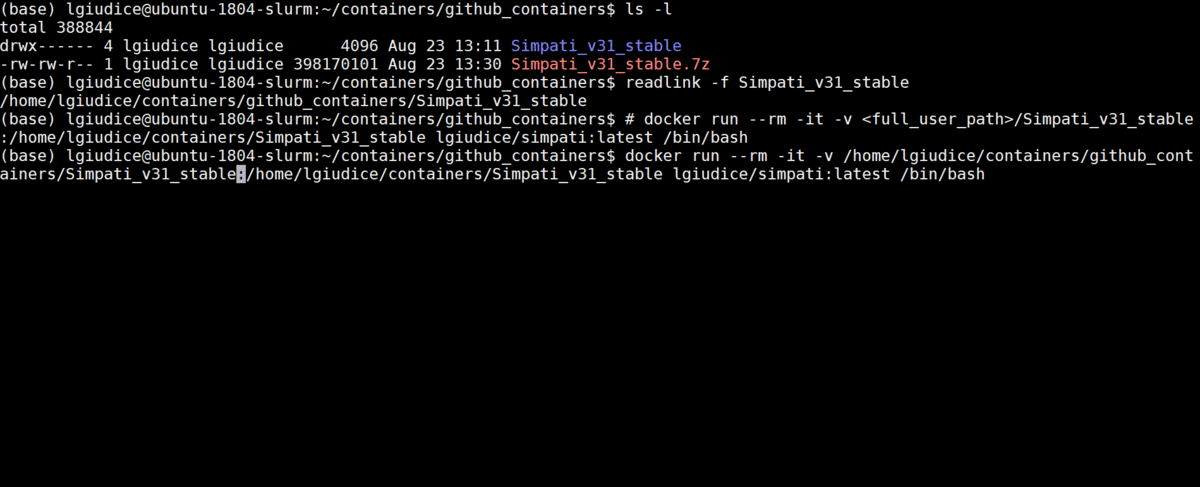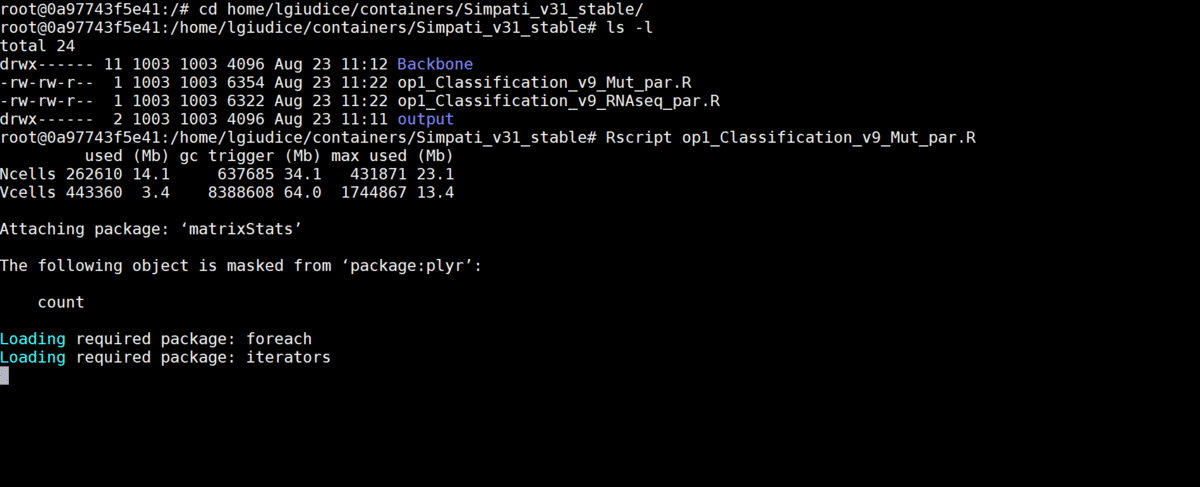
- Download and unzip the Simpati data and scripts from the link
- With bash:
wget https://univr-my.sharepoint.com/:u:/g/personal/luca_giudice_univr_it/EeQJQb335QlGtTdMd3ni2mIB79wUeVy151SpVG8ULWqi2w?download=1 --no-check-certificate -O Simpati_v31_stable.7z
- With bash:
- Simpati_v31_stable/output/op1 directory contains the results used in the publication, if you want to get fresh new output data you can remove it
- Run the simpati container building the unzipped Simpati directory to the following path: /home/lgiudice/containers/Simpati_v31_stable
- With bash:
docker run --rm -it -v <full_user_path>/Simpati_v31_stable:/home/lgiudice/containers/Simpati_v31_stable lgiudice/simpati:latest /bin/bash
- With bash:
- Modify the script op1_Classification_v9_Mut_par.R at the line 91 and 96 based on your computational resources, setting the RAM memory and the number of cores
- Modify the script op1_Classification_v9_RNAseq_par.R at the line 91 and 96 based on your computational resources, setting the RAM memory and the number of cores
- Now you are ready to run Simpati:
Rscript op1_Classification_v9_Mut_par.R Rscript op1_Classification_v9_RNAseq_par.R
- Download and unzip the netDx data and scripts from the link
- With bash:
wget https://univr-my.sharepoint.com/:u:/g/personal/luca_giudice_univr_it/EdeJn9yyrfFNv0rAikZF1gYBdoLzM_9HGWU8Pyw7wVJNhQ?download=1 --no-check-certificate -O netDx_Mut.7z
- With bash:
- netDx/output directory contains the results used in the publication, if you want to get fresh new output data you can remove everything except: op2_TCGA4netDx_mut
- Run the comp1 container building the unzipped netDx directory to the following path: /home/lgiudice/containers/netDx_Mut
- With bash:
docker run --rm -it -v <full_user_path>/netDx_Mut:/home/lgiudice/containers/netDx_Mut lgiudice/comp1:latest /bin/bash
- With bash:
- Modify the script op3_multi_omics_TCGA.R at the line 101 based on your computational resources, setting the RAM memory and the number of cores
- Modify the script op3_multi_omics_TCGA_large.R at the line 101 based on your computational resources, setting the RAM memory and the number of cores
- Now you are ready to run netDx:
Rscript op3_multi_omics_TCGA.R Rscript op3_multi_omics_TCGA_large.R
- Download and unzip the netDx data and scripts from the following link
- With bash:
wget https://univr-my.sharepoint.com/:u:/g/personal/luca_giudice_univr_it/EXaGdLX9_e5Huj_KjoOfF4gB_HcOou9ghoTQ1zF0ZI77zw?download=1 --no-check-certificate -O netDx_RNAseq.7z
- With bash:
- netDx/output directory contains the results used in the publication, if you want to get fresh new output data you can remove everything except: op2_TCGA4netDx_RNAseq
- Run the comp1 container building the unzipped netDx directory to the following path: /home/lgiudice/containers/netDx_RNAseq
- With bash:
docker run --rm -it -v <full_user_path>/netDx_RNAseq:/home/lgiudice/containers/netDx_RNAseq lgiudice/comp1:latest /bin/bash
- With bash:
- Modify the script op3_multi_omics_TCGA.R at the line 68 based on your computational resources, setting the RAM memory and the number of cores
- Modify the script op3_multi_omics_TCGA_large.R at the line 68 based on your computational resources, setting the RAM memory and the number of cores
- Now you are ready to run netDx
Rscript op3_multi_omics_TCGA.R Rscript op3_multi_omics_TCGA_large.R
- Download and unzip the PASNet data and scripts from the following link
- PASNet/output//output_py directory contains the results used in the publication for a specific dataset, if you want to get fresh new output data you can delete that directory
- Run the comp2 container building the unzipped PASNet directory to the following path: /home/PASNet
- With bash:
docker run --rm -it -v <full_user_path>/PASNet:/home/PASNet lgiudice/comp2:latest /bin/bash
- With bash:
- Now you are ready to run PASNet workflow: first you will tune the model and then run the model with the tuned parameters
python op1_Run_EmpiricalSearch python op2_Run