-
Notifications
You must be signed in to change notification settings - Fork 18
obtaining marker genes
To run read2tree two things are required as input:
- The DNA sequencing reads as FASTQ file(s).
- A set of reference orthologous groups, i.e. marker genes.
We provide two sets of markergenes for Mammalia and Bacteria here. These could be used along with the arguments --standalone_path marker_genes/ --dna_reference {bacteria/mammalia}_cnda.fa . However, we recommend using OMA browser to download set of marker genes tailored for the clade of interest to accurately infer the species tree.
In this page we describe how to obtain the latter using OMA browser.
Step 1. Open the Export gene marker genes page of OMA browser using this link.

This page can be found on the Download tab of the main page of OMA browser as well.

Step 2. In the field search by species name, you can type the name of species or a clade, e.g. primates. Then click on the found item in blue.
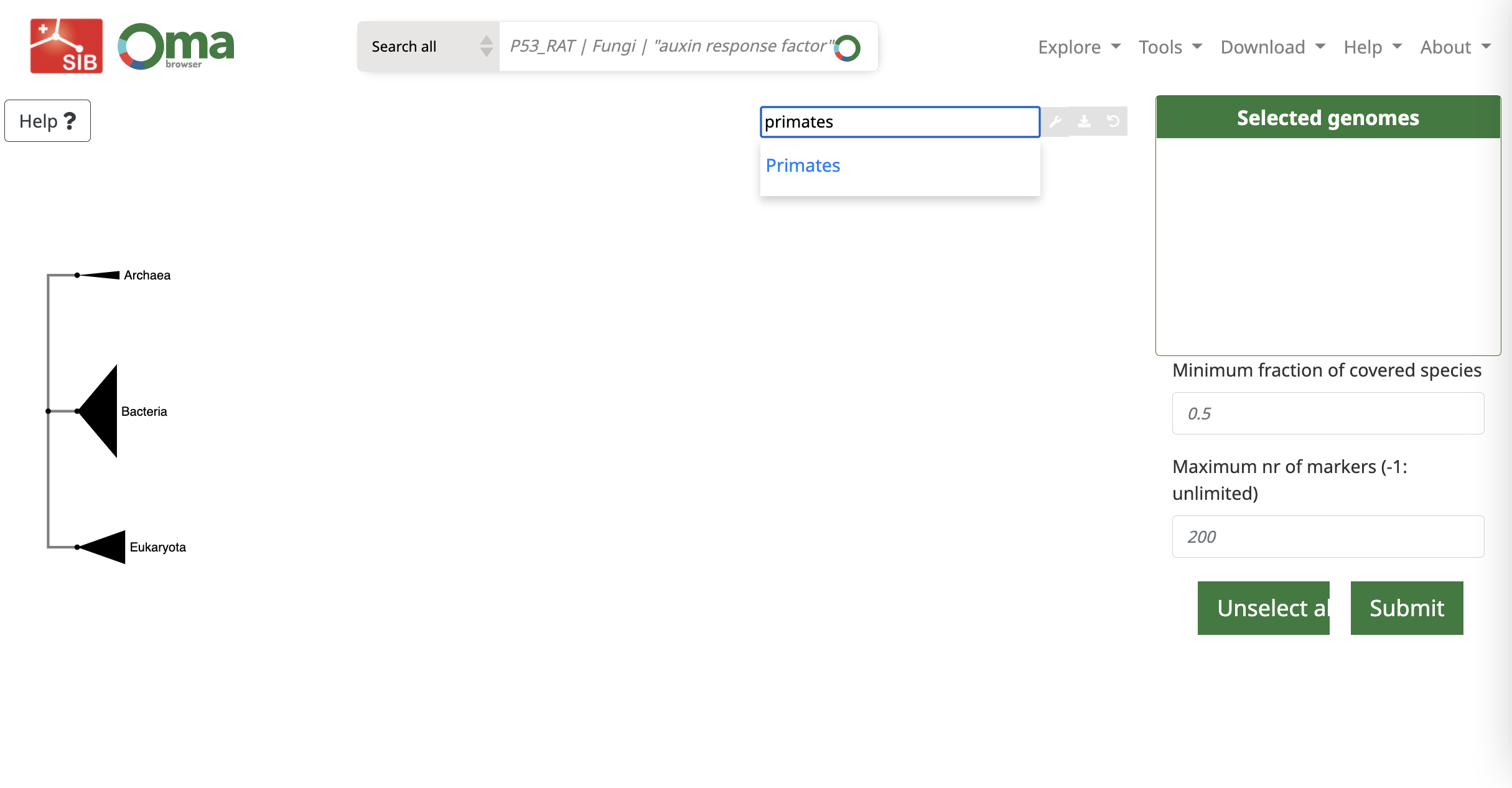
Step 3. Then click on the internal node or leaves of the tree of life, and select (all) species. You can also expand or collapse the nodes.

Step 4. Now on the right side, we can set the value of Minimum fraction of covered species to 0.8 and Maximum nr of markers to 500 (for studying a small clade 100 is probably enough). However, if you are not limited in computation, you can set this to -1 to prepare all the possible OGs.

Then, click submit

Step 5. Finally, after waiting for few mins/hours (depending how large is your species set), a compressed file containing the FASTA files of the marker genes is ready to download.


Then you need to combine the fna files
tar xvzf marker_genes_*.tgz
ls marker_genes/*.fna | wc -l
cat marker_genes/*.fna > dna_ref.fa
Note: If you want to infer the tree for viruses, check this instruction. In summary, we recommend to first download a set of proteomes and their CNDA from NCBI refSeq for your clade of interest, then use [OMA standalone] (https://omabrowser.org/standalone/) to infer the set of marker genes.
For corona virus, you can use this link to download marker genes in addition to download the cdna fasta file (1MB) from here and unzip it. Once you have the marker genes as proteome and cnda, you can use the sequencing reads of your samples to infer the species tree all together using read2tree using the argument --standalone_path marker_genes --dna_reference viruses.cdna.fa .