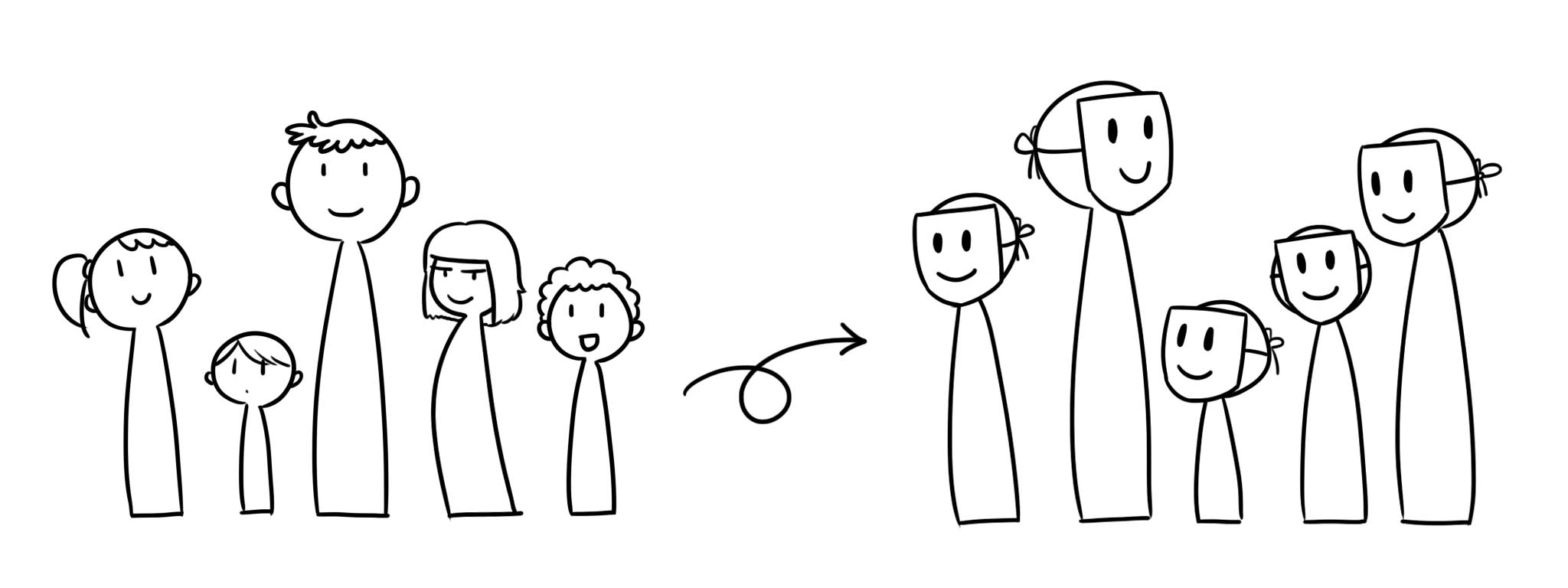
twinify is a software package for privacy-preserving generation of a synthetic twin to a given sensitive tabular data set.
On a high level, twinify follows the differentially private data sharing process introduced by Jälkö et al.. Depending on the nature of your data, twinify implements either the NAPSU-MQ approach described by Räisä et al. or finds an approximate parameter posterior for any probabilistic model you formulated using differentially private variational inference (DPVI). For the latter, twinify also offers automatic modelling for easy building of models fitting the data. If you have existing experience with NumPyro you can also implement your own model directly.
Often data that would be very useful for the scientific community is subject to privacy regulations and concerns and cannot be shared. Differentially private data sharing allows to generate synthetic data that is statistically similar to the original data - the synthetic twin - while at the same time satisfying a mathematical privacy formulation known as differential privacy. Differential privacy measures the level of privacy in terms of positive parameters ε and δ - where smaller values imply stronger privacy - thus giving us concrete knobs to tune the synthetic data generation to our privacy needs and ensuring that private information remains private!
In order to generate data, we rely on probabilistic modelling, which means we assume the data follows a probability distribution with some parameters which we can infer privately. In order to generate the synthetic twin data, we sample from this distribution with the learned parameters, the posterior predictive distribution.
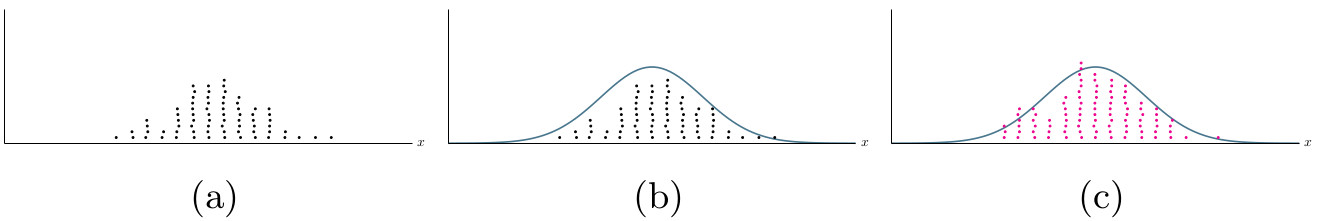
As an example, consider a population of individuals with varying height shown in the first panel of the illustrations above. We present the heights as a histogram in panel (a) of the figure below. We then fit a probabilistic model for this data, the blue curve in in panel (b), and sample new data from this distribution, the magenta dots in (c).
As the learning of the model is performed under differential privacy, the sampled data preserves the anonymity of individuals while maintaining the statistical properties of the original population. This is shown in the second panel in the illustration above.
twinify can be used as a software library from your own application or as a stand-alone command line tool operating on data sets provided as a CSV file. Either way, the high-level steps are the same and we outline them in the following for the command line tool. You can find a brief overview of twinify's API for library use further below. Please also check the software documentation for more detailed information.
The first thing you need to do is decide whether you want to use the NAPSU-MQ approach or learn a probabilistic model using DPVI. NAPSU-MQ
- NAPSU-MQ learns a maximum entropy distribution that best reproduces a user-chosen set of marginal queries on the data. NAPSU-MQ produces a model that encapsulates the additional uncertainty introduced by differential privacy. However, currently it is only suitable for fully categorical data. May exhibit long runtimes for data sets with many feature dimensions.
- DPVI is capable of learning any probabilistic model you specify, for categorical, continuous or mixed data. However, the result is only an approximation to the true posterior and it is unable to explicitly capture additional uncertainty due to differential privacy.
If you have fully categorical data, you will likely obtain better results with NAPSU-MQ. However, if your data has a large number of feature dimensions, you may find that you can get acceptable results in shorter time using DPVI.
If your data contains non-categorical features, DPVI is your only choice without resorting to discretization. DPVI might also be an interesting option if you have strong data-independent prior knowledge that you want to incorporate into your model.
The main thing you need to do next for either method is to define the probabilistic model to be learned. The following describes the modelling approaches for the different methods, assuming an input csv file with three features that are titled Age and Height (cm) and Eye color.
For NAPSU-MQ this means that you must specify the the marginal queries to preserve. You can in principle select any number of queries with any subset of features, however, the larger the number of queries, the longer the fitting of the model will take.
To specify marginal queries, you have to create a text file in which you list one query per line and all features covered by the query using the corresonding column name in the data csv file, separated by commas.
We assume here that the features Age and Height are discretized and require NAPSU-MQ to fit all feature marginals as well as the two-way marginal over the combined features Age and Height, resulting in the following model/query file:
Age
Height (cm)
Age, Height (cm)
Eye color
twinifys automatic modelling feature for DPVI builds a mixture model for user specified feature distributions. Technically speaking, the feature distribution specifies the distribution of the feature conditioned on the latent mixture component assignment. Under this conditioning, feature distributions are assumed to be independent.
To specify the feature distributions, you have to create a text file in which you only need to specify a single distribution for each of your features. For the assumed example the model file might look like:
Age : Poisson
# you can also have comments in here
Height (cm): Normal
Eye color : Categorical
A example of such text file for a larger data set is available in examples/covid19_analysis/models/full_model.txt. In automatic modelling twinify chooses a suitable non-/weakly informative prior for the parameters of the feature distribution. It also automates the encoding of string valued features into a suitable domain according to the chosen feature distribution.
If you are familiar with the NumPyro probabilistic programming framework and want a more flexible way of specifying models, you can provide a Python file containing NumPyro code to twinify. All you need to do is define a model function that specifies the NumPyro model for a single data instance x. You also have to define functions for pre- and postprocessing of data (if required). You can find details on the exact requirements for NumPyro models in the FAQ below and an example in examples/covid19_analysis/models/numpyro_model_example.py.
Once you have have set the probabilistic model, you can run twinify by calling from your command line
twinify [napsu|vi] input_data_path model_path output_path_prefix
where the model is specified as
- NAPSU-MQ: text file containing marginal queries
- DPVI: either the text file for automatic modelling or as a python module that contains the NumPyro model.
twinify will output the generated synthetic data as output_path_prefix.csv and a file with learned model parameters as output_path_prefix.p.
There are a number of (optional) command line arguments that further influence twinify's behaviour:
-
--epsilon- Privacy parameter ε (positive real number): Use this argument to specify the ε privacy level. Smaller is better (but may negatively impact utility). In general values less than 1 are considered strong privacy and values less than 2 still reasonable. -
--delta- Privacy parameter δ (positive real number between 0 and 1): Use this argument to override the default choice for δ (should rarely be required). Smaller is better. Recommended to be less than 1/N, where N is the size of your data set. Values larger are typically considered unsafe. -
--num_synthetic- Number of synthetic samples (integer): Use this to set how many samples you want from the generative model. This has no effect on the privacy guarantees for the synthetic data. -
--seed- Stochasticity seed (integer): Use this argument to seed the initial random state to fix internal stochasticity of twinify if you need reproducibility. twinify will use a strong source of randomness by default if this argument is not given. -
--drop_na- Preprocessing behavior: Use this flag to remove any data instances with at least one missing value.
Command line arguments specific to DPVI (ignored by NAPSU-MQ):
--k- Number of mixture components (integer): Use this argument to set the number of mixture components when automatic modelling is used. A reasonable choice would be of same magnitude as the number of features.--sampling_ratio,-q- Subsampling ratio (real number between 0 and 1): Use this argument to set the relative size of subsets (batches) of data the iteratively private learning is uses. This has privacy implications and is further discussed in FAQ.--num_epochs,-e, - Number of learning epochs (integer): Use this argument to set the number of passes through the data (epochs) the private learning performs. This has privacy implications and is further discussed in FAQ.--clipping_threshold- Privacy parameter (positive real number): Use this argument to adapt the clipping of gradients, an internal parameter for the private learning that limits how much each sample can effect the learning. It is only advised for experienced users to change this parameter.
As an example, say we have data in my_data.csv and a model description for DPVI with automatic modelling in my_model.txt. We want 1000 samples of generated data to be stored in my_twin.csv and fix twinify's internal randomness with a seed for reproducibility. This is how we run twinify:
twinify vi my_data.csv my_model.txt my_twin --seed=123 --num_synthetic=1000
In the case that we wrote a model with NumPyro instead of relying on twinify's automatic modelling, our call would like like
twinify vi my_data.csv my_numpyro_model.py my_twin --seed=123 --num_synthetic=1000
Assuming that the data is entirely categorical and that we have set up a list of marginal queries in my_queries.txt, we can run twinify using NAPSU-MQ with the following command:
twinify napsu my_data.csv my_queries.txt my_twin --seed=123 --num_synthetic=1000
Using twinify as a library, you retain full control over data loading, pre- and postprocessing, in contrast to the command line tool.
The main actors in the twinify APIs are twinify.InferenceModel and twinify.InferenceResult.
InferenceModel fully encapsulates a model and algorithm to fit it to the data. It defines a single function
fit(data: pd.DataFrame, rng: d3p.random.PRNGState, epsilon: float, delta: float, **kwargs) -> InferenceResult
which takes an input data set given as a pandas DataFrame as well as privacy parameters and a randomness state. It returns
a representation of the model fitted to the data in the form of a InferenceResult object.
Currently twinify provides twinify.dpvi.DPVIModel and twinify.napsu_mq.NapsuMQModel as concrete implementations, with the following initializers:
DPVIModel(model: NumPyroModelFunction, guide: Optional[NumPyroGuideFunction] = None, clipping_threshold: float = 1., num_epochs: int = 1000, subsample_ratio: float = 0.01)NapsuMQModel(required_marginals: Iterable[FrozenSet[str]] = tuple(), use_laplace_approximation: bool = True)
InferenceResult represents a learned model from which synthetic data can be generated. To that end it defines the method
generate(
rng: d3p.random.PRNGState,
num_parameter_samples: int,
num_data_per_parameter_sample: int = 1,
single_dataframe: bool = True
) -> Union[Iterable[pd.DataFrame], pd.DataFrame]
This method first draws num_parameter_samples parameter samples from the model posterior represented by the InferenceResult object and then
samples num_data_per_parameter_sample data points for each parameter sample from the model, and returns them as either one large combined DataFrame or an iterable over one DataFrame per parameter sample.
InferenceResult classes also allow saving and loading of learned models via the save and static load methods respectively.
Note that DPVIResult.load requires the same NumPyro model as used for inference to be provided during model loading.
You can check out a short example of how to use twinify as a library implementing the NAPSU-MQ approach through twinify.napsu_mq.NapsuMQModel and
twinify.napsu_mq.NapsuMQResult in the jupyter notebook in examples/NapsuMQ example.ipynb.
A stable version of twinify can be installed from the Python Package Index via pip using the following command:
pip install twinify
Alternatively, you can install twinify from the cloned repository to get the current development version (this might contain breaking changes, however):
git clone https://github.com/DPBayes/twinify
cd twinify
pip install .
twinify relies on NumPyro, a versatile probabilistic programming framework similar to Pyro, for modelling and inference purposes. NumPyro uses fast CPU and GPU kernels for execution, which are provided by the JAX framework. Differentially private training routines for NumPyro are introduced by the d3p package.
First off, we need to warn you about tweaking the hyperparameters based on quality of the synthetic data: If you do that your choice will end up tailored to your specific data set which can leak private information in subtle ways, degrading the privacy guarantees given by twinify. Unfortunately, there's is no simple way to work around that other than finding good parameters on a similar public data set before working on your sensitive data.
If it is possible, you can usually improve quality of the synthetic data by relaxing your privacy constraints (i.e., choosing a larger ε for the same δ).
Also, differentially private learning is known to work better with more data. In case you are working with particularly small data set, you might need to collect more data in order to improve the utility of synthetic data.
Real data is often incomplete and missing values might occur for a multitude of reasons, for example due to scarcity in measuring resources. twinify supports modelling features with missing values using a simple mechanism: It assumes that values can be missing at random (independently from whether other feature values are missing as well) with a certain probability that is inferred from the data. During data generation, twinify first evaluates whether there should be a value, and, if so, samples one from the feature distribution specified in the model you provided.
Using automatic modelling, twinify detects and handles features with missing values automatically and you don't need to do anything. You can disable that behavior by setting the --drop_na=1 command line argument to remove all data instances with missing values.
When writing your own NumPyro models, you can use the twinify.na_model.NAModel class to wrap around the feature distribution for achieving the same effect.
In mathematical terms, the likelihood of data in the NAModel is
where is the likelihood of existing data x (according to the assigned feature distribution) and
denotes the probability that x is missing. Similar to other model parameters, twinify assigns a prior to and learns a posterior for
.
Currently supported feature distributions are shown in the table below with the corresponding prior choices twinify uses for the parameters of these distributions.
| Distribution | Parameters | Priors | Use for |
|---|---|---|---|
| Normal | location μ, scale σ | μ ∼ 𝓝(0, 10),σ ∼ LogNormal(0,2) | (symmetric) continuous real numbers |
| Bernoulli | probability p | p ∼ Beta(1, 1) | binary categories (0/1 integers or "yes"/"no" strings) |
| Categorical | probabilities p | p ∼ Dirichlet(1, ..., 1) | arbitrary categories (integer or string data) |
| Poisson | rate λ | λ ∼ Exp(1) | ordinal integer data |
As already mentioned, twinify's automatic modelling uses the distributions you specify for each feature (i.e., column in the data) to build a so called mixture model consisting of several components. In each mixture component, the features are assumed to be independently modelled by the distributions you specified with component-specific parameters. Each data instance is associated with a single component with a probability given by the mixture's weight. During data generation, for each generated data instance, twinify first randomly picks a component according to the weights and then samples the data point according from the parameterised feature distributions in that component.
While all features are treated as independent in each mixture component, the mixture model as a whole is typically able to capture correlations between features.
In mathematical terms, the likelihood of the data given the model parameters for the mixture model is
where is the density function of the user-defined feature distribution and
is the d-th feature column of the data set. To complete the probabilistic model twinify assigns non-informative prior distributions to the model parameters
as well as the weights
for each of the K mixture components.
There are only a few constraints twinify imposes. These are listed below.
You must define a function model(x = None, num_obs_total = None) containing the NumPyro model with the following constraints:
modelhandles a single data instance at once and gets all data features in a single vector, i.e.,xhas shape(num_features,).- Feature values in
xare ordered as they appear in the data set. num_obs_totalis the number of total observations (i.e., the size of your data set) that you can use to scale the likelihood accordingly.- During data generation,
xandnum_obs_totalwill both beNone. modelmust return a sample forxwith features following the same order as in the input.
You may specify a SVI guide function with the same arguments as models. If you do not, twinify uses NumPyro's automatic guides.
You may specify a preprocessing function preprocess(loaded_data) that gets the data as a pandas.DataFrame as it was loaded from the csv-file and returns a data frame which rows will be passed to model during inference. Your preprocessing may involve
- selecting the relevant feature columns out of the data
- reordering feature columns
- mapping strings to numeric values
- etc.
If you do not specify a preprocessing function, no preprocessing will take place and the loaded data is used as is.
You may specify a post-processing function postprocess(sampled_data) that gets the data as a pandas.DataFrame sampled from the model after inference and returns a data frame to be written to the output csv-file. A possible post-processing step would be to map numeric values back to their string representation (i.e., reversing the mapping applied during preprocessing). If you do not specify a post-processing function, no post-processing will take place the generated data is stored as is.
The private learning algorithm twinify uses is based on gradient descent optimization using perturbed gradients in every iteration. The gradients are first clipped so that their norm does not exceed a given threshold and then perturbed using Gaussian noise to mask any individuals contribution. Larger variance of Gaussian noise leads to more strict privacy guarantees, i.e., to smaller ε and δ.
twinify accepts the privacy level ε (and δ, typically determined automatically) as parameters and finds the variance for Gaussian noise to suffice this level of privacy. The noise variance is additionally affected by the number of epochs (Nₑ) and the subsampling ratio (q) as σ² ~= O(q Nₑ) since both affect the number of total iterations the algorithm performs and thus the number of times private data is handled.
Larger noise variance can negatively affect the learning so choosing too large values for q or Nₑ will likely give bad results.
twinify version numbers adhere to Semantic Versioning. Changes
between releases are tracked in ChangeLog.txt.
twinify's code base is licensed under the Apache License 2.0.
Some files of the accompanying documentation and examples may be licensed differently. You can find an annotation
about which license applies in the beginning of each file using SPDX tags (or in a separate file named <file>.license for files where
this information cannot be directly embedded).
The full license texts can be founds in the LICENSES/ folder in this directory.
When using twinify, please cite
@article{jalko19,
title={Privacy-preserving data sharing via probabilistic modelling},
author={Joonas Jälkö and Eemil Lagerspetz and Jari Haukka and Sasu Tarkoma and Samuel Kaski and Antti Honkela},
year={2021},
journal={Patterns},
volume={2},
number={7},
publisher={Elsevier}
}
For the NAPSU-MQ method, cite
@inproceedings{raisa23,
title={Noise-Aware Statistical Inference with Differentially Private Synthetic Data},
author={Räisä, Ossi and Jälkö, Joonas and Kaski, Samuel and Honkela, Antti},
booktitle={Proceedings of the 26th International Conference on Artificial Intelligence and Statististics},
year={2023},
pages={3620--3643},
volume={206},
series={Proceedings of Machine Learning Research}
}