-
Notifications
You must be signed in to change notification settings - Fork 6
Home
The following details the steps involved in:
- Generating a Trinity de novo RNA-Seq assembly
- Evaluating the quality of the assembly
- Quantifying transcript expression levels
- Identifying differentially expressed (DE) transcripts
- Functionally annotating transcripts using Trinotate and predicting coding regions using TransDecoder
- Examining functional enrichments for DE transcripts using GOseq
- Interactively Exploring annotations and expression data via TrinotateWeb
All required software and data are provided pre-installed on an Amazon EC2 AMI.
The workshop materials here expect that you have a minimal familiarity with UNIX.
After launching and connecting to your running instance of the AMI, change your working directory to the base directory of these workshop materials. We'll get there by moving down three directories:
% cd workshop_materials
% cd transcriptomics
% cd KrumlovTrinityWorkshopJan2018
Yes, you could have just 'cd workshop_materials/transcriptomics/KrumlovTrinityWorkshopJan2018', but here we're taking the slower scenic route. Feel free to look around and run 'ls' to see what other files or directories are there along the way and get a feel for where you are in the file system - which is particularly useful if you're new to unix.
This 'KrumlovTrinityWorkshopJan2018' will be the base working directory for all the exercises below. We'll create other subdirectories here and move back and forth to our various workspaces that we generate along the way.
Below, we refer to '%' as the terminal command prompt, and we use environmental variables such as $TRINITY_HOME and $TRINOTATE_HOME as shortcuts to referring to their installation directories in the AMI image. To view the path to the installation directories, you can simply run:
% echo $TRINITY_HOME
Also, some commands can be fairly long to type in, an so they'll be more easily displayed in this document, we separate parts of the command with '\' characters and put the rest of the command on the following line. Similarly in unix you can type '[return]' and you can continue to type in the rest of the command on the new line in the terminal.
For viewing text files, we'll use the unix utilities 'head' (look at the top few lines), 'cat' (print the entire contents of the file), and 'less' (interactively page through the file), and for viewing PDF formatted files, we'll use the 'xpdf' viewer utility.
Before we begin, let's slightly update our PATH setting to ensure certain Trinity-plugins will be found properly.
% export PATH=$TRINITY_HOME/trinity-plugins/BIN/:${PATH}
For this course we will be using the data from this paper: Defining the transcriptomic landscape of Candida glabrata by RNA-Seq. Linde et al. Nucleic Acids Res. 2015 This work provides a detailed RNA-Seq-based analysis of the transcriptomic landscape of C. glabrata in nutrient-rich media (WT), as well as under nitrosative stress (GSNO), in addition to other conditions, but we'll restrict ourselves to just WT and GSNO conditions for demonstration purposes in this workshop.
There are paired-end FASTQ formatted Illlumina read files for each of the two conditions, with three biological replicates for each. All RNA-Seq data sets can be found in the data/ subdirectory:
% ls -1 data | grep fastq
.
GSNO_SRR1582646_1.fastq
GSNO_SRR1582646_2.fastq
GSNO_SRR1582647_1.fastq
GSNO_SRR1582647_2.fastq
GSNO_SRR1582648_1.fastq
GSNO_SRR1582648_2.fastq
wt_SRR1582649_1.fastq
wt_SRR1582649_2.fastq
wt_SRR1582650_1.fastq
wt_SRR1582650_2.fastq
wt_SRR1582651_1.fastq
wt_SRR1582651_2.fastq
Each biological replicate (eg. wt_SRR1582651) contains a pair of fastq files (eg. wt_SRR1582651_1.fastq.gz for the 'left' and wt_SRR1582651_2.fastq.gz for the 'right' read of the paired end sequences). Normally, each file would contain millions of reads, but in order to reduce running times as part of the workshop, each file provided here is restricted to only 10k RNA-Seq reads.
It's generally good to evaluate the quality of your input data using a tool such as FASTQC. Since exploration of FASTQC reports has already been done in a previous section of this workshop, we'll skip doing it again here - and trust that the quality of these reads meet expectations.
Finally, another set of files that you will find in the data include 'mini_sprot.pep*', corresponding to a highly abridged version of the SWISSPROT database, containing only the subset of protein sequences that are needed for use in this workshop. It's provided and used here only to speed up certain operations, such as BLAST searches, which will be performed at several steps in the tutorial below. Of course, in exploring your own RNA-Seq data, you would leverage the full version of SWISSPROT and not this tiny subset used here.
To generate a reference assembly that we can later use for analyzing differential expression, we'll combine the read data sets for the different conditions together into a single target for Trinity assembly. We do this by providing Trinity with a list of the targeted fastq files organized according to sample type and replicate name, as provided in a 'samples.txt' file.
Take a look at the samples.txt file:
% cat data/samples.txt
.
GSNO GSNO_SRR1582646 data/GSNO_SRR1582646_1.fastq data/GSNO_SRR1582646_2.fastq
GSNO GSNO_SRR1582647 data/GSNO_SRR1582647_1.fastq data/GSNO_SRR1582647_2.fastq
GSNO GSNO_SRR1582648 data/GSNO_SRR1582648_1.fastq data/GSNO_SRR1582648_2.fastq
wt wt_SRR1582649 data/wt_SRR1582649_1.fastq data/wt_SRR1582649_2.fastq
wt wt_SRR1582650 data/wt_SRR1582650_1.fastq data/wt_SRR1582650_2.fastq
wt wt_SRR1582651 data/wt_SRR1582651_1.fastq data/wt_SRR1582651_2.fastq
Using this samples.txt file, perform de novo transcriptome assembly of the reads with Trinity like so:
% ${TRINITY_HOME}/Trinity --seqType fq --samples_file data/samples.txt \
--CPU 2 --max_memory 2G --min_contig_length 150
Note, if you see a message about not being able to identify the version of Java, please just ignore it.
Running Trinity on this data set may take 10 to 15 minutes. You'll see it progress through the various stages, starting with Jellyfish to generate the k-mer catalog, then followed by Inchworm to assemble 'draft' contigs, Chrysalis to cluster the contigs and build de Bruijn graphs, and finally Butterfly for tracing paths through the graphs and reconstructing the final isoform sequences.
Running a typical Trinity job requires ~1 hour and ~1G RAM per ~1 million PE reads. You'd normally run it on a high-memory machine and let it churn for hours or days.
The assembled transcripts will be found at 'trinity_out_dir/Trinity.fasta'.
Just to look at the top few lines of the assembled transcript fasta file, you can run:
% head trinity_out_dir/Trinity.fasta
and you can see the Fasta-formatted Trinity output:
>TRINITY_DN506_c0_g1_i1 len=171 path=[149:0-170] [-1, 149, -2]
TGAGTATGGTTTTGCCGGTTTGGCTGTTGGTGCAGCTTTGAAGGGCCTAAAGCCAATTGT
TGAATTCATGTCATTCAACTTCTCCATGCAAGCCATTGACCATGTCGTTAACTCGGCAGC
AAAGACACATTATATGTCTGGTGGTACCCAAAAATGTCAAATCGTGTTCAG
>TRINITY_DN512_c0_g1_i1 len=168 path=[291:0-167] [-1, 291, -2]
ATATCAGCATTAGACAAAAGATTGTAAAGGATGGCATTAGGTGGTCGAAGTTTCAGGTCT
AAGAAACAGCAACTAGCATATGACAGGAGTTTTGCAGGCCGGTATCAGAAATTGCTGAGT
AAGAACCCATTCATATTCTTTGGACTCCCGTTTTGTGGAATGGTGGTG
>TRINITY_DN538_c0_g1_i1 len=310 path=[575:0-309] [-1, 575, -2]
GTTTTCCTCTGCGATCAAATCGTCAAACCTTAGACCTAGCTTGCGGTAACCAGAGTACTT
Note, the sequences you see will likely be different, as the order of sequences in the output is not deterministic.
The FASTA sequence header for each of the transcripts contains the identifier for the transcript (eg. 'TRINITY_DN506_c0_g1_i1'), the length of the transcript, and then some information about how the path was reconstructed by the software by traversing nodes within the graph.
It is often the case that multiple isoforms will be reconstructed for the same 'gene'. Here, the 'gene' identifier corresponds to the prefix of the transcript identifier, such as 'TRINITY_DN506_c0_g1', and the different isoforms for that 'gene' will contain different isoform numbers in the suffix of the identifier (eg. TRINITY_DN506_c0_g1_i1 and TRINITY_DN506_c0_g1_i2 would be two different isoform sequences reconstructed for the single gene TRINITY_DN506_c0_g1). It is useful to perform certain downstream analyses, such as differential expression, at both the 'gene' and at the 'isoform' level, as we'll do later below.
There are several ways to quantitatively as well as qualitatively assess the overall quality of the assembly, and we outline many of these methods at our Trinity wiki.
You can count the number of assembled transcripts by using 'grep' to retrieve only the FASTA header lines and piping that output into 'wc' (word count utility) with the '-l' parameter to just count the number of lines.
% grep '>' trinity_out_dir/Trinity.fasta | wc -l
How many were assembled?
It's useful to know how many transcript contigs were assembled, but it's not very informative. The deeper you sequence, the more transcript contigs you will be able to reconstruct. It's not unusual to assemble over a million transcript contigs with very deep data sets and complex transcriptomes, but as you 'll see below (in the section containing the more informative guide to assembly assessment) a fraction of the transcripts generally best represent the input RNA-Seq reads.
Capture some basic statistics about the Trinity assembly:
% $TRINITY_HOME/util/TrinityStats.pl trinity_out_dir/Trinity.fasta
which should generate data like so. Note your numbers may vary slightly, as the assembly results are not deterministic.
################################
## Counts of transcripts, etc.
################################
Total trinity 'genes': 683
Total trinity transcripts: 687
Percent GC: 44.39
########################################
Stats based on ALL transcript contigs:
########################################
Contig N10: 742
Contig N20: 525
Contig N30: 423
Contig N40: 346
Contig N50: 300
Median contig length: 216
Average contig: 279.85
Total assembled bases: 192257
#####################################################
## Stats based on ONLY LONGEST ISOFORM per 'GENE':
#####################################################
Contig N10: 728
Contig N20: 524
Contig N30: 420
Contig N40: 343
Contig N50: 296
Median contig length: 215
Average contig: 278.14
Total assembled bases: 189969
The total number of reconstructed transcripts should match up identically to what we counted earlier with our simple 'grep | wc' command. The total number of 'genes' is also reported - and simply involves counting up the number of unique transcript identifier prefixes (without the _i isoform numbers). When the 'gene' and 'transcript' identifiers differ, it's due to transcripts being reported as alternative isoforms for the same gene. In our tiny example data set, we reconstruct only a small number of alternative isoforms, and note that alternative splicing in this yeast species may be fairly rare. Tackling an insect or mammal transcriptome would be expected to yield many alternative isoforms.
You'll also see 'Contig N50' values reported. You'll remmeber from the earlier lectures on genome assembly that the 'N50 statistic indicates that at least half of the assembled bases are in contigs of at least that contig length'. We extend the N50 statistic to provide N40, N30, etc. statistics with similar meaning. As the N-value is decreased, the corresponding length will increase.
Most of this is not quantitatively useful, and the values are only reported for historical reasons - it's simply what everyone used to do in the early days of transcriptome assembly. The N50 statistic in RNA-Seq assembly can be easily biased in the following ways:
-
Overzealous reconstruction of long alternatively spliced isoforms: If an assembler tends to generate many different 'versions' of splicing for a gene, such as in a combinatorial way, and those isoforms tend to have long sequence lengths, the N50 value will be skewed towards a higher value.
-
Highly sensitive reconstruction of lowly expressed isoforms: If an assembler is able to reconstruct transcript contigs for those transcirpts that are very lowly expressed, these contigs will tend to be short and numerous, biasing the N50 value towards lower values. As one sequences deeper, there will be more evidence (reads) available to enable reconstruction of these short lowly expressed transcripts, and so deeper sequencing can also provide a downward skew of the N50 value.
We now move into the section containing more meaningful metrics for evaluating your transcriptome assembly.
A high quality transcriptome assembly is expected to have strong representation of the reads input to the assembler. By aligning the RNA-Seq reads back to the transcriptome assembly, we can quantify read representation. Use the Bowtie2 aligner to align the reads to the Trinity assembly, and in doing so, take notice of the read representation statistics reported by the bowtie2 aligner.
First build a bowtie2 index for the Trinity assembly, required before running the alignment:
% bowtie2-build trinity_out_dir/Trinity.fasta trinity_out_dir/Trinity.fasta
Now, align the reads to the assembly:
% bowtie2 --local --no-unal -x trinity_out_dir/Trinity.fasta \
-q -1 data/wt_SRR1582651_1.fastq -2 data/wt_SRR1582651_2.fastq \
| samtools view -Sb - | samtools sort -o - - > bowtie2.bam
.
[bam_header_read] EOF marker is absent. The input is probably truncated.
[samopen] SAM header is present: 686 sequences.
10000 reads; of these:
10000 (100.00%) were paired; of these:
6922 (69.22%) aligned concordantly 0 times
2922 (29.22%) aligned concordantly exactly 1 time
156 (1.56%) aligned concordantly >1 times
----
6922 pairs aligned concordantly 0 times; of these:
191 (2.76%) aligned discordantly 1 time
----
6731 pairs aligned 0 times concordantly or discordantly; of these:
13462 mates make up the pairs; of these:
12476 (92.68%) aligned 0 times
752 (5.59%) aligned exactly 1 time
234 (1.74%) aligned >1 times
37.62% overall alignment rate
Generally, in a high quality assembly, you would expect to see at least ~70% aligned and at least ~70% of the reads to exist as proper pairs. Our tiny read set used here in this workshop does not provide us with a high quality assembly, as only ~30% of aligned reads are mapped as proper pairs - which is usually the sign of a fractured assembly. In this case, deeper sequencing and assembly of more reads would be expected to lead to major improvements here.
Every assembled transcript is only as valid as the reads that support it. If you ever want to examine the read support for one of your favorite transcripts, you could do this using the IGV browser.
Let's examine the above bowtie2 alignments to our Trinity transcripts using IGV. Before viewing the bam file, we must first index it using samtools:
% samtools index bowtie2.bam
Then, you can launch IGV on these data like so:
% igv.sh -g trinity_out_dir/Trinity.fasta bowtie2.bam
Note, you could also launch IGV via clicking the icon on the desktop and then manually loading in the various input files via the menu, but that does take some time. Launching it from the command line is rather straightforward and fast.
Select different transcripts for viewing. Since we only aligned one of our sets of reads, you may not find coverage across entire sequences. We would need to align all the reads for a more comprehensive view. We'll skip that for now.

Take some time to familiarize yourself with IGV. Look at a few transcripts and consider the read support. View the reads as pairs to examine the paired-read linkages. (hint: cntrl-click, 'view as pairs').
Another very useful metric in evaluating your assembly is to assess the number of fully reconstructed coding transcripts. This can be done by performing a BLASTX search of your assembled transcript sequences to a high quality database of protein sequences, such as provided by SWISSPROT. Searching a large protein database using BLASTX can take a while - longer than we want during this workshop, so instead, we'll search the mini-version of SWISSPROT that comes installed in our data/ directory:
% blastx -query trinity_out_dir/Trinity.fasta \
-db data/mini_sprot.pep -out blastx.outfmt6 \
-evalue 1e-20 -num_threads 2 -max_target_seqs 1 -outfmt 6
The above blastx command will have generated an output file 'blastx.outfmt6', storing only the single best matching protein given the E-value threshold of 1e-20.
Examine the formatting of the tab-delimited blast output file:
% head blastx.outfmt6 | column -t
.
TRINITY_DN1_c0_g1_i1 YP010_YEAST 70.18 57 16 1 73 240 1 57 6e-24 83.2
TRINITY_DN10_c0_g1_i1 RL21A_YEAST 93.88 147 9 0 442 2 1 147 4e-100 283
TRINITY_DN11_c0_g1_i1 RS24B_YEAST 95.90 122 5 0 367 2 1 122 7e-69 202
TRINITY_DN14_c0_g1_i1 VATB_YEAST 96.49 57 2 0 3 173 430 486 2e-33 114
TRINITY_DN14_c0_g2_i1 VATB_YEAST 95.60 91 4 0 3 275 328 418 1e-56 179
TRINITY_DN15_c0_g1_i1 RS15_YEAST 80.39 51 9 1 18 170 1 50 9e-24 83.6
TRINITY_DN15_c0_g2_i1 RS15_YEAST 87.88 99 12 0 3 299 44 142 2e-61 182
TRINITY_DN16_c0_g1_i1 CISY1_YEAST 90.38 343 33 0 2 1030 136 478 0.0 640
TRINITY_DN16_c0_g2_i1 CISY1_YEAST 70.09 107 29 1 322 2 26 129 4e-46 151
TRINITY_DN18_c0_g1_i1 ALF_YEAST 88.14 59 7 0 2 178 268 326 6e-33 111
Can you figure out which columns correspond to the Trinity transcript, it's matching database sequence, the percent identity, and the E-value for match significance?
By running another script in the Trinity suite, we can compute the length representation of best matching SWISSPROT matches like so:
% $TRINITY_HOME/util/analyze_blastPlus_topHit_coverage.pl \
blastx.outfmt6 trinity_out_dir/Trinity.fasta \
data/mini_sprot.pep | column -t
.
#hit_pct_cov_bin count_in_bin >bin_below
100 78 78
90 18 96
80 11 107
70 19 126
60 15 141
50 24 165
40 33 198
30 40 238
20 62 300
10 24 324
The above table lists bins of percent length coverage of the best matching protein sequence along with counts of proteins found within that bin. For example, 78 proteins are matched by more than 90% of their length up to 100% of their length. There are 18 matched by more than 80% and up to 90% of their length. The third column provides a running total, indicating that 96 transcripts match more than 80% of their length, and 107 transcripts match more than 70% of their length, etc.
The count of full-length transcripts is going to be dependent on how good the assembly is in addition to the depth of sequencing, but should saturate at higher levels of sequencing. Performing this full-length transcript analysis using assemblies at different read depths and plotting the number of full-length transcripts as a function of sequencing depth will give you an idea of whether or not you've sequenced deeply enough or you should consider doing more RNA-Seq to capture more transcripts and obtain a better (more complete) assembly.
We'll explore some additional metrics that are useful in assessing the assembly quality below, but they require that we estimate expression values for our transcripts, so we'll tackle that first.
To estimate transcript expression values, we'll use the salmon software. We'll run salmon on each of the sample replicates as listed in our samples.txt file:
% $TRINITY_HOME/util/align_and_estimate_abundance.pl --seqType fq \
--samples_file data/samples.txt --transcripts trinity_out_dir/Trinity.fasta \
--est_method salmon --trinity_mode --prep_reference --output_dir salmon
The above should have generated separate sets of outputs for each of the sample replicates. Examine the new contents of your working directory:
% ls -ltr
.
...
drwxr-xr-x 9 bhaas 1594166068 306 Jan 5 18:19 wt_SRR1582651
drwxr-xr-x 9 bhaas 1594166068 306 Jan 5 18:21 GSNO_SRR1582646
drwxr-xr-x 9 bhaas 1594166068 306 Jan 5 18:21 GSNO_SRR1582648
drwxr-xr-x 9 bhaas 1594166068 306 Jan 5 18:21 GSNO_SRR1582647
drwxr-xr-x 9 bhaas 1594166068 306 Jan 5 18:21 wt_SRR1582650
drwxr-xr-x 9 bhaas 1594166068 306 Jan 5 18:21 wt_SRR1582649
Take a look at the contents of one of these salmon output directories:
% ls -ltr wt_SRR1582651
.
drwxr-xr-x 3 bhaas 1594166068 102 Jan 5 18:19 logs
drwxr-xr-x 3 bhaas 1594166068 102 Jan 5 18:19 libParams
drwxr-xr-x 8 bhaas 1594166068 272 Jan 5 18:19 aux_info
-rw-r--r-- 1 bhaas 1594166068 30752 Jan 5 18:21 quant.sf.genes
-rw-r--r-- 1 bhaas 1594166068 30181 Jan 5 18:21 quant.sf
-rw-r--r-- 1 bhaas 1594166068 631 Jan 5 18:21 lib_format_counts.json
-rw-r--r-- 1 bhaas 1594166068 432 Jan 5 18:21 cmd_info.json
Examine the contents of the 'quant.sf' file:
% head wt_SRR1582651/quant.sf | column -t
.
Name Length EffectiveLength TPM NumReads
TRINITY_DN0_c0_g1_i1 308 157.95 1965.28 9
TRINITY_DN1_c0_g1_i1 240 96.0038 2155.59 6
TRINITY_DN10_c0_g1_i1 473 319.649 539.512 5
TRINITY_DN11_c0_g1_i1 416 262.791 1181.23 9
TRINITY_DN12_c0_g1_i1 362 209.731 1151.17 7
TRINITY_DN14_c0_g1_i1 174 43.8077 787.324 1
TRINITY_DN14_c0_g2_i1 277 128.996 534.759 2
TRINITY_DN15_c0_g1_i1 172 42.4787 811.956 1
TRINITY_DN15_c0_g2_i1 309 158.847 434.264 2
The key columns in the above salmon output are the transcript identifier 'Name', the 'NumReads' corresponding to the number of RNA-Seq fragments predicted to be derived from that transcript, and the 'TPM' column indicates the normalized expression values for the expression of that transcript in the sample (measured as Transcripts Per Million).
Now, given the expression estimates for each of the transcripts in each of the samples, we're going to pull together all values into matrices containing transcript IDs in the rows, and sample names in the columns. We'll make two matrices, one containing the estimated counts, and another containing the TPM expression values that are cross-sample normalized using the TMM method. This is all done for you by the following script in Trinity, indicating the method we used for expresssion estimation and providing the list of individual sample abundance estimate files.
First, let's create a list of the quant.sf files:
% find wt_* GSNO_* -name "quant.sf" | tee quant_files.list
.
wt_SRR1582649/quant.sf
wt_SRR1582650/quant.sf
wt_SRR1582651/quant.sf
GSNO_SRR1582646/quant.sf
GSNO_SRR1582647/quant.sf
GSNO_SRR1582648/quant.sf
Using this new file 'quant_files.list', we'll use a Trinity script to generate the count and expression matrices for both the transcript isoforms and sepearate files for 'gene's.
% $TRINITY_HOME/util/abundance_estimates_to_matrix.pl --est_method salmon \
--out_prefix Trinity --name_sample_by_basedir \
--quant_files quant_files.list \
--gene_trans_map trinity_out_dir/Trinity.fasta.gene_trans_map
You should find a matrix file called 'Trinity.isoform.counts.matrix', which contains the counts of RNA-Seq fragments mapped to each transcript.
Examine the first few lines of the counts matrix:
% head -n20 Trinity.isoform.counts.matrix | column -t
.
wt_SRR1582649 wt_SRR1582650 wt_SRR1582651 GSNO_SRR1582646 GSNO_SRR1582647 GSNO_SRR1582648
TRINITY_DN543_c0_g1_i1 0 4 1 1 2 0
TRINITY_DN256_c0_g3_i1 13 5 8 0 1 0
TRINITY_DN288_c0_g3_i1 29 20 22 0 0 0
TRINITY_DN596_c0_g1_i1 1 1 1 2 3 0
TRINITY_DN353_c0_g1_i1 3 0 0 1 1 2
TRINITY_DN260_c0_g2_i1 0 0 1 2 6 2
TRINITY_DN235_c0_g1_i2 3 0 4 8 15 9.30823
TRINITY_DN276_c0_g2_i1 26 17 30 2 4 2
TRINITY_DN527_c0_g1_i1 4 4 4 28 34 29
You'll see that the above matrix has integer values representing the number of RNA-Seq paired-end fragments that are estimated to have been derived from that corresponding transcript in each of the samples. Don't be surprised if you see some values that are not exact integers but rather fractional read counts. This happens if there are multiply-mapped reads (such as to common sequence regions of different isoforms), in which case the multiply-mapped reads are fractionally assigned to the corresponding transcripts according to their maximum likelihood.
The counts matrix will be used by DESeq2 (or other tools in Bioconductor) for statistical analysis and identifying significantly differentially expressed transcripts.
Now take a look at the first few lines of the normalized expression matrix:
% head -n20 Trinity.isoform.TMM.EXPR.matrix | column -t
.
wt_SRR1582649 wt_SRR1582650 wt_SRR1582651 GSNO_SRR1582646 GSNO_SRR1582647 GSNO_SRR1582648
TRINITY_DN543_c0_g1_i1 0.000 4285.916 1207.919 1250.354 2318.497 0.000
TRINITY_DN256_c0_g3_i1 2882.375 1075.231 1763.201 0.000 219.023 0.000
TRINITY_DN288_c0_g3_i1 2429.634 1634.889 1688.699 0.000 0.000 0.000
TRINITY_DN596_c0_g1_i1 1083.186 1009.491 1133.029 2358.738 3293.404 0.000
TRINITY_DN353_c0_g1_i1 2738.546 0.000 0.000 994.050 930.488 2204.007
TRINITY_DN260_c0_g2_i1 0.000 0.000 721.127 1501.711 4235.108 1677.503
TRINITY_DN235_c0_g1_i2 365.070 0.000 457.735 980.110 1779.601 1347.575
TRINITY_DN276_c0_g2_i1 1690.132 1078.912 1764.242 129.311 251.891 154.163
TRINITY_DN527_c0_g1_i1 313.156 305.603 285.846 2186.494 2582.453 2694.495
These are the normalized expression values, which have been further cross-sample normalized using TMM normalization to adjust for any differences in sample composition. TMM normalization assumes that most transcripts are not differentially expressed, and linearly scales the expression values of samples to better enforce this property. TMM normalization is described in A scaling normalization method for differential expression analysis of RNA-Seq data, Robinson and Oshlack, Genome Biology 2010.
We use the TMM-normalized expression matrix when plotting expression values in heatmaps and other expression analyses.
Note, similar count and expression files were generated at the 'gene' level as well, and these can be used similarly to the isoform matrices wherever you want to perform a gene-based analysis instead. It's often useful to study the expression data at both the gene and isoform level, particularly in cases where differential transcript usage exists (isoform switching), where differences in expression may not be apparent at the gene level.
Although we outline above several of the reasons for why the contig N50 statistic is not a useful metric of assembly quality, below we describe the use of an alternative statistic - the ExN50 value, which we assert is more useful in assessing the quality of the transcriptome assembly. The ExN50 indicates the N50 contig statistic (as earlier) but restricted to the top most highly expressed transcripts. Compute it like so:
% $TRINITY_HOME/util/misc/contig_ExN50_statistic.pl Trinity.isoform.TMM.EXPR.matrix \
trinity_out_dir/Trinity.fasta > ExN50.stats
View the contents of the above output file:
% cat ExN50.stats | column -t
Note, your results may vary slightly.
.
E ExN50 num_transcripts
1 329 1
2 329 2
3 247 3
4 247 4
5 329 5
6 329 7
7 329 9
8 327 11
9 303 13
10 327 15
11 329 17
....
35 415 101
36 417 105
37 416 110
38 417 115
39 420 121
40 417 126
41 417 131
42 420 137
43 420 143
44 417 148
45 416 154
....
69 330 326
70 329 334
71 329 343
72 328 352
73 325 361
74 328 371
75 325 380
76 320 389
77 320 399
78 320 409
79 320 419
....
89 303 530
90 305 542
91 303 555
92 302 569
93 303 582
94 303 597
95 292 612
96 296 628
97 302 644
98 302 662
99 302 685
100 302 686
The above table indicates the contig N50 value based on the entire transcriptome assembly (E100), which is a small value (285). By restricting the N50 computation to the set of transcripts representing the top most 65% of expression data, we obtain an N50 value of 420.
Try plotting the ExN50 statistics:
% $TRINITY_HOME/util/misc/plot_ExN50_statistic.Rscript ExN50.stats
% xpdf ExN50.stats.plot.pdf

As you can see, the N50 value will tend to peak at a value higher than that computed using the entire data set. With a high quality transcriptome assembly, the N50 value should peak at ~90% of the expression data, which we refer to as the E90N50 value. Reporting the E90N50 contig length and the E90 transcript count are more meaningful than reporting statistics based on the entire set of assembled transcripts. Remember the caveat in assembling this tiny data set. A plot based on a larger set of reads looks like so:
Assembly using 900K PE reads:

Assembly using 4.5M PE reads:
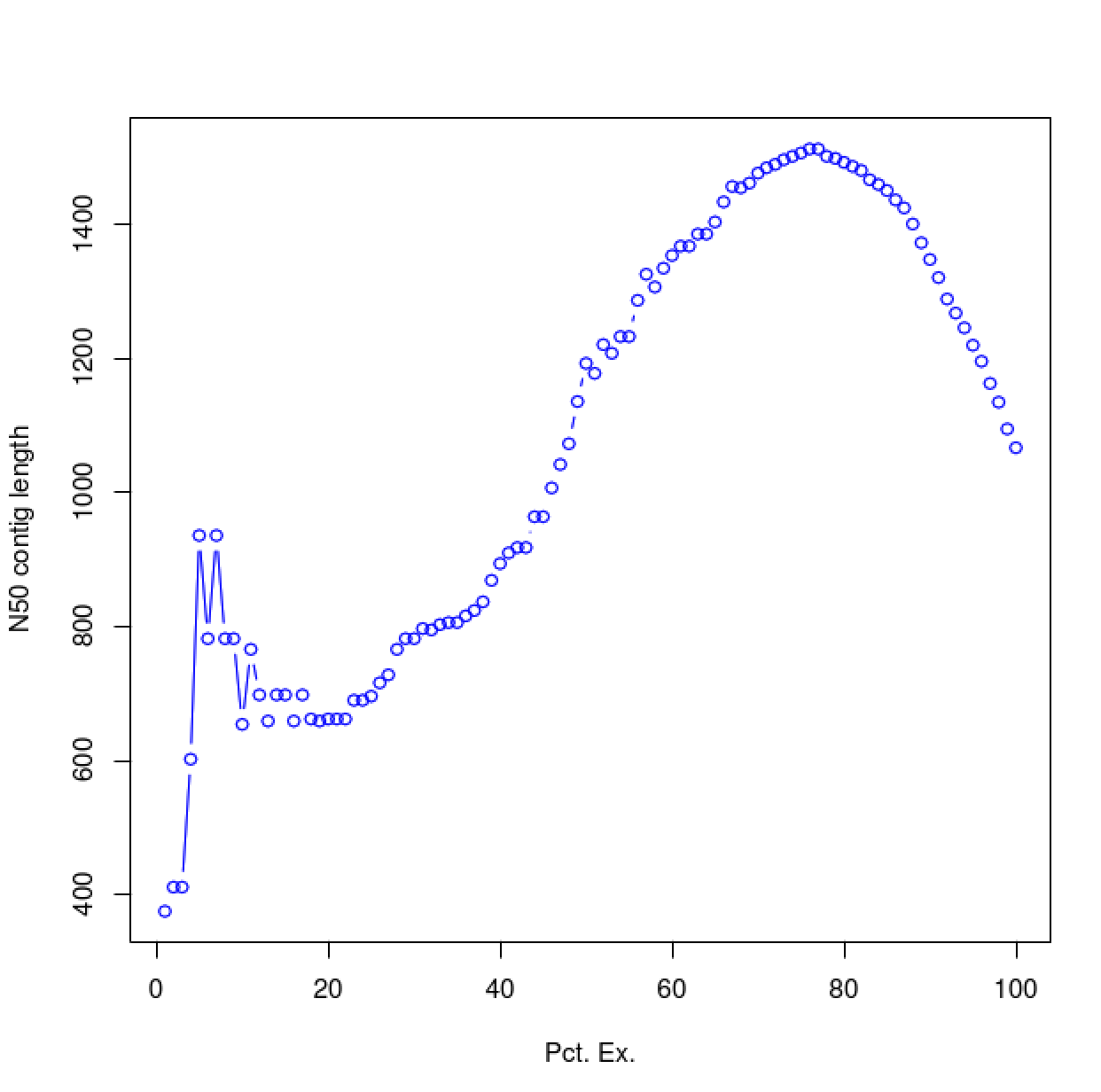
Assembly using 18M PE reads:
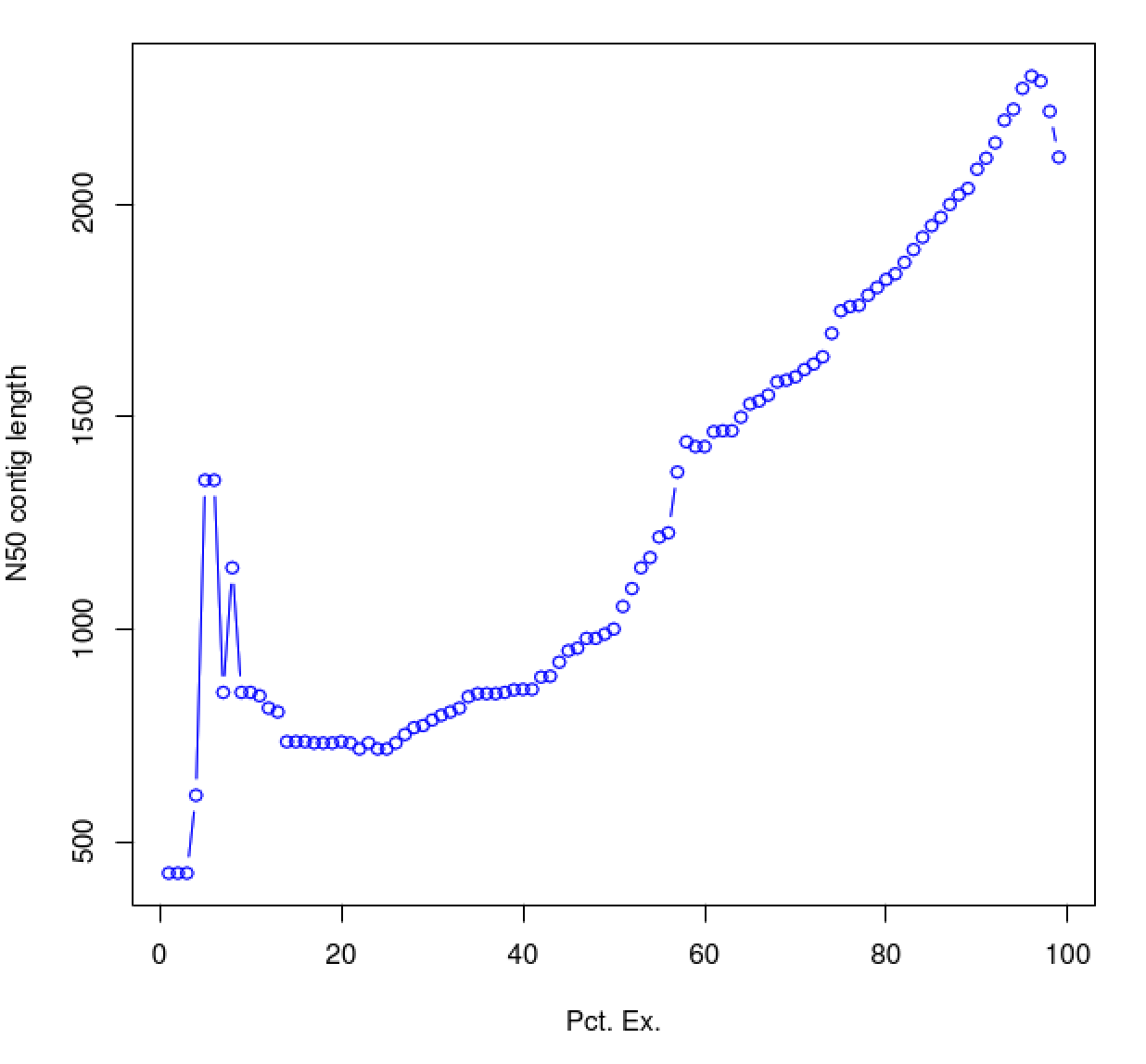
You can see that as you sequence deeper, you'll end up with an assembly that has an ExN50 peak that approaches the use of ~90% of the expression data.
A plethora of tools are currently available for identifying differentially expressed transcripts based on RNA-Seq data, and of these, DESeq2 is among the most popular and most accurate. The DESeq2 software is part of the R Bioconductor package, and we provide support for using it in the Trinity package.
Having biological replicates for each of your samples is crucial for accurate detection of differentially expressed transcripts. In our data set, we have three biological replicates for each of our conditions, and in general, having three or more replicates for each experimental condition is highly recommended.
To detect differentially expressed transcripts, run the Bioconductor package DESeq2 using our counts matrix:
% $TRINITY_HOME/Analysis/DifferentialExpression/run_DE_analysis.pl \
--matrix Trinity.isoform.counts.matrix \
--samples_file data/samples.txt \
--method DESeq2 \
--output DESeq2_trans
Examine the contents of the DESeq2_trans/ directory.
% ls -ltr DESeq2_trans/
.
-rw-r--r-- 1 bhaas 1594166068 1545 Jan 5 23:56 Trinity.isoform.counts.matrix.GSNO_vs_wt.DESeq2.Rscript
-rw-r--r-- 1 bhaas 1594166068 24550 Jan 5 23:57 Trinity.isoform.counts.matrix.GSNO_vs_wt.DESeq2.count_matrix
-rw-r--r-- 1 bhaas 1594166068 15522 Jan 5 23:57 Trinity.isoform.counts.matrix.GSNO_vs_wt.DESeq2.DE_results.MA_n_Volcano.pdf
-rw-r--r-- 1 bhaas 1594166068 115612 Jan 5 23:57 Trinity.isoform.counts.matrix.GSNO_vs_wt.DESeq2.DE_results
The files '*.DE_results' contain the output from running DESeq2 to identify differentially expressed transcripts in each of the pairwise sample comparisons. Examine the format of one of the files, such as the results from comparing Sp_log to Sp_plat:
% head DESeq2_trans/Trinity.isoform.counts.matrix.GSNO_vs_wt.DESeq2.DE_results | column -t
.
sampleA sampleB baseMeanA baseMeanB baseMean log2FoldChange lfcSE stat pvalue padj
TRINITY_DN486_c0_g1_i1 GSNO wt 16.8981745355167 106.980365419029 61.9392699772726 -2.6493893326134 0.255388975436606 -10.3739377476419 3.25815666078611e-25 1.7384467428526e-22
TRINITY_DN577_c0_g1_i1 GSNO wt 15.7288302868206 101.644075065183 58.6864526760018 -2.69899406362077 0.261493095561904 -10.3214735280911 5.63515962026775e-25 1.7384467428526e-22
TRINITY_DN556_c0_g1_i1 GSNO wt 23.9663919729509 105.796343729641 64.8813678512961 -2.15116963920305 0.233191735883499 -9.2248965472677 2.83834758240544e-20 5.8375348611472e-18
TRINITY_DN324_c0_g1_i1 GSNO wt 1.47231222746358 80.2499964184142 40.8611543229389 -5.79076278854971 0.677003202157668 -8.55352348422289 1.19386992588915e-17 1.84154436068402e-15
TRINITY_DN310_c0_g1_i1 GSNO wt 1.93665704962814 64.4895090163414 33.2130830329848 -4.99435027412214 0.588931323085661 -8.48036108515101 2.24496126493146e-17 2.77028220092542e-15
TRINITY_DN157_c0_g2_i1 GSNO wt 53.21625596387 4.41363265791558 28.8149443108928 3.59667509622987 0.4743359674083 7.58254769479438 3.38834460951473e-14 3.48434770678431e-12
TRINITY_DN142_c0_g1_i1 GSNO wt 0 64.3364882003275 32.1682441001638 -8.65066949183313 1.19947555120541 -7.21204319932964 5.5118480566932e-13 4.55603207727768e-11
TRINITY_DN143_c0_g1_i1 GSNO wt 1.10944933305853 52.0143601526098 26.5619047428341 -5.49448910934864 0.762847590122535 -7.20260400700233 5.90733494622713e-13 4.55603207727768e-11
TRINITY_DN601_c0_g1_i1 GSNO wt 71.5561235105584 16.9551033864134 44.2556134484859 2.06065562273661 0.293266276985126 7.02656863216878 2.11674484549853e-12 1.45114618852511e-10
These data include the log fold change (logFC), log counts per million (logCPM), P- value from an exact test, and false discovery rate (FDR).
The DESeq2 analysis above generated both MA and Volcano plots based on these data. Examine any of these like so:
% xpdf DESeq2_trans/Trinity.isoform.counts.matrix.GSNO_vs_wt.DESeq2.DE_results.MA_n_Volcano.pdf

The red data points correspond to all those features that were identified as being significant with an FDR <= 0.05.
Exit the chart viewer to continue.
Trinity facilitates analysis of these data, including scripts for extracting transcripts that are above some statistical significance (FDR threshold) and fold-change in expression, and generating figures such as heatmaps and other useful plots, as described below.
Now let's perform the following operations from within the DESeq2_trans/ directory. Enter the DESeq2_trans/ dir like so:
% cd DESeq2_trans/
Extract those differentially expressed (DE) transcripts that are at least 4-fold differentially expressed at a significance of <= 0.001 in any of the pairwise sample comparisons:
% $TRINITY_HOME/Analysis/DifferentialExpression/analyze_diff_expr.pl \
--matrix ../Trinity.isoform.TMM.EXPR.matrix \
--samples ../data/samples.txt \
-P 1e-3 -C 2
The above generates several output files with a prefix diffExpr.P1e-3_C2', indicating the parameters chosen for filtering, where P (FDR actually) is set to 0.001, and fold change (C) is set to 2^(2) or 4-fold. (These are default parameters for the above script. See script usage before applying to your data).
Included among these files are: ‘diffExpr.P1e-3_C2.matrix’ : the subset of the FPKM matrix corresponding to the DE transcripts identified at this threshold. The number of DE transcripts identified at the specified thresholds can be obtained by examining the number of lines in this file.
% wc -l diffExpr.P1e-3_C2.matrix
.
(n) diffExpr.P1e-3_C2.matrix
where n ~ 100 to 110
Note, the number of lines in this file includes the top line with column names, so there are actually (n-1) DE transcripts at this 4-fold and 1e-3 FDR threshold cutoff.
Also included among these files is a heatmap 'diffExpr.P1e-3_C2.matrix.log2.centered.genes_vs_samples_heatmap.pdf' as shown below, with transcripts clustered along the vertical axis and samples clustered along the horizontal axis.
% xpdf diffExpr.P1e-3_C2.matrix.log2.centered.genes_vs_samples_heatmap.pdf

The expression values are plotted in log2 space and mean-centered (mean expression value for each feature is subtracted from each of its expression values in that row), and shows upregulated expression as yellow and downregulated expression as purple.
Exit the PDF viewer to continue.
Extract clusters of transcripts with similar expression profiles by cutting the transcript cluster dendrogram at a given percent of its height (ex. 60%), like so:
% $TRINITY_HOME/Analysis/DifferentialExpression/define_clusters_by_cutting_tree.pl \
--Ptree 60 -R diffExpr.P1e-3_C2.matrix.RData
This creates a directory containing the individual transcript clusters, including a pdf file that summarizes expression values for each cluster according to individual charts:
% xpdf diffExpr.P1e-3_C2.matrix.RData.clusters_fixed_P_60/my_cluster_plots.pdf

You can do all the same analyses as you did above at the gene level. For now, let's just rerun the DE detection step, since we'll need the results later on for use with TrinotateWeb. Also, it doesn't help us to study the 'gene' level data with this tiny data set (yet another disclaimer) given that all our transcripts = genes, since we didn't find any alternative splicing variants. With typical data sets, you will have alterantively spliced isoforms identified, and performing DE analysis at the gene level should provide more power for detection than at the isoform level. For more info about this, I encourage you to read this paper.
Before running the gene-level DE analysis, be sure to back out of the current DESeq2_trans/ directory like so:
% cd ../
Be sure you're in your base working directory:
% pwd
.
/home/genomics/workshop_materials/transcriptomics/KrumlovTrinityWorkshopJan2018
Now, run the DE analysis at the gene level like so:
% $TRINITY_HOME/Analysis/DifferentialExpression/run_DE_analysis.pl \
--matrix Trinity.gene.counts.matrix \
--samples_file data/samples.txt \
--method DESeq2 \
--output DESeq2_gene
You'll now notice that the DESeq2_gene/ directory exists and is populated with similar files.
% ls -ltr DESeq2_gene/
Let's move on and make use of those outputs later. With your own data, however, you would normally run the same set of operations as you did above for the transcript-level DE analyses.
Now we have a bunch of transcript sequences and have identified some subset of them that appear to be biologically interesting in that they're differentially expressed between our two conditions - but we don't really know what they are or what biological functions they might represent. We can explore their potential functions by functionally annotating them using our Trinotate software and analysis protocol. To learn more about Trinotate, you can visit the Trinotate website.
Again, let's make sure that we're back in our primary working directory called 'KrumlovTrinityWorkshopJan2018':
% pwd
.
/home/genomics/workshop_materials/transcriptomics/KrumlovTrinityWorkshopJan2018
If you're not in the above directory, then relocate yourself to it.
Now, create a Trinotate/ directory and relocate to it. We'll use this as our Trinotate computation workspace.
% mkdir Trinotate
% cd Trinotate
Below, we're going to run a number of different tools to capture information about our transcript sequences.
TransDecoder is a tool we built to identify likely coding regions within transcript sequences. It identifies long open reading frames (ORFs) within transcripts and scores them according to their sequence composition. Those ORFs that encode sequences with compositional properties (codon frequencies) consistent with coding transcripts are reported.
Running TransDecoder is a two-step process. First run the TransDecoder step that identifies all long ORFs.
% $TRANSDECODER_HOME/TransDecoder.LongOrfs -t ../trinity_out_dir/Trinity.fasta
Now, run the step that predicts which ORFs are likely to be coding.
% $TRANSDECODER_HOME/TransDecoder.Predict -t ../trinity_out_dir/Trinity.fasta
You'll now find a number of output files containing 'transdecoder' in their name:
% ls -1 |grep transdecoder
.
Trinity.fasta.transdecoder.bed
Trinity.fasta.transdecoder.cds
Trinity.fasta.transdecoder.gff3
Trinity.fasta.transdecoder.pep
Trinity.fasta.transdecoder_dir/
...
The file we care about the most here is the 'Trinity.fasta.transdecoder.pep' file, which contains the protein sequences corresponding to the predicted coding regions within the transcripts.
Go ahead and take a look at this file:
% less Trinity.fasta.transdecoder.pep
.
>TRINITY_DN107_c0_g1_i1.p1 TRINITY_DN107_c0_g1~~TRINITY_DN107_c0_g1_i1.p1 ORF type:internal len:175 (+),score=164.12 TRINITY_DN107_c0_g1_i1:2-523(+)
VPLYQHLADLSDSKTSPFVLPVPFLNVLNGGSHAGGALALQEFMIAPTGAKSFREAMRIG
SEVYHNLKSLTKKRYGSSAGNVGDEGGVAPDIQTAEEALDLIVDAIKAAGHEGKVKIGLD
CASSEFFKDGKYDLDFKNPNSDASKWLSGPQLADLYHSLVKKYPIVSIEDPFAE
>TRINITY_DN10_c0_g1_i1.p2 TRINITY_DN10_c0_g1~~TRINITY_DN10_c0_g1_i1.p2 ORF type:internal len:158 (-),score=122.60 TRINITY_DN10_c0_g1_i1:2-472(-)
TDQDKRYQAKMGKSHGYRSRTRYMFQRDFRKHGAIALSTYLKVYKVGDIVDIKANGSIQK
GMPHKFYQGKTGVVYNVTKSSVGVIVNKMVGNRYLEKRLNLRVEHVKHSKCRQEFLDRVK
SNAAKRAEAKAQGKAVQLKRQPAQPREARVVSTEGNV
>TRINITY_DN110_c0_g1_i1.p2 TRINITY_DN110_c0_g1~~TRINITY_DN110_c0_g1_i1.p2 ORF type:complete len:131 (+),score=98.69 TRINITY_DN110_c0_g1_i1:55-447(+)
MTRSSVLADALNAINNAEKTGKRQVLIRPSSKVIIKFLQVMQRHGYIGEFEYIDDHRSGK
Type 'q' to exit the 'less' viewer.
There are a few items to take notice of in the above peptide file. The header lines includes the protein identifier composed of the original transcripts along with '|m.(number)'. The 'type' attribute indicates whether the protein is 'complete', containing a start and a stop codon; '5prime_partial', meaning it's missing a start codon and presumably part of the N-terminus; '3prime_partial', meaning it's missing the stop codon and presumably part of the C-terminus; or 'internal', meaning it's both 5prime-partial and 3prime-partial. You'll also see an indicator (+) or (-) to indicate which strand the coding region is found on, along with the coordinates of the ORF in that transcript sequence.
This .pep file will be used for various sequence homology and other bioinformatics analyses below.
Earlier, we ran blastx against our mini SWISSPROT datbase to identify likely full-length transcripts. Let's run blastx again to capture likely homolog information, and we'll lower our E-value threshold to 1e-5 to be less stringent than earlier.
% blastx -db ../data/mini_sprot.pep \
-query ../trinity_out_dir/Trinity.fasta -num_threads 2 \
-max_target_seqs 1 -outfmt 6 -evalue 1e-5 \
> swissprot.blastx.outfmt6
Now, let's look for sequence homologies by just searching our predicted protein sequences rather than using the entire transcript as a target:
% blastp -query Trinity.fasta.transdecoder.pep \
-db ../data/mini_sprot.pep -num_threads 2 \
-max_target_seqs 1 -outfmt 6 -evalue 1e-5 \
> swissprot.blastp.outfmt6
Using our predicted protein sequences, let's also run a HMMER search against the Pfam database, and identify conserved domains that might be indicative or suggestive of function:
% hmmscan --cpu 2 --domtblout TrinotatePFAM.out \
../data/trinotate_data/Pfam-A.hmm \
Trinity.fasta.transdecoder.pep
Note, hmmscan might take a few minutes to run.
The signalP and tmhmm software tools are very useful for predicting signal peptides (secretion signals) and transmembrane domains, respectively.
To predict signal peptides, run signalP like so:
% signalp -f short -n signalp.out Trinity.fasta.transdecoder.pep
Take a look at the output file:
% less signalp.out
.
##gff-version 2
##sequence-name source feature start end score N/A ?
## -----------------------------------------------------------
TRINITY_DN19_c0_g1_i1|m.141 SignalP-4.0 SIGNAL 1 18 0.553 . . YES
TRINITY_DN33_c0_g1_i1|m.174 SignalP-4.0 SIGNAL 1 19 0.631 . . YES
....
How many of your proteins are predicted to encode signal peptides?
Generating a Trinotate annotation report involves first loading all of our bioinformatics computational results into a Trinotate SQLite database. The Trinotate software provides a boilerplate SQLite database called 'Trinotate.sqlite' that comes pre-populated with a lot of generic data about SWISSPROT records and Pfam domains (and is a pretty large file consuming several hundred MB). Below, we'll populate this database with all of our bioinformatics computes and our expression data.
As a sanity check, be sure you're currently located in your 'Trinotate/' working directory.
% pwd
.
/home/genomics/workshop_materials/transcriptomics/KrumlovTrinityWorkshopJan2018/Trinotate
Copy the provided Trinotate.sqlite boilerplate database into your Trinotate working directory like so:
% cp ../data/trinotate_data/Trinotate.boilerplate.sqlite Trinotate.sqlite
Load your Trinotate.sqlite database with your Trinity transcripts and predicted protein sequences:
% $TRINOTATE_HOME/Trinotate Trinotate.sqlite init \
--gene_trans_map ../trinity_out_dir/Trinity.fasta.gene_trans_map \
--transcript_fasta ../trinity_out_dir/Trinity.fasta \
--transdecoder_pep Trinity.fasta.transdecoder.pep
Load in the various outputs generated earlier:
% $TRINOTATE_HOME/Trinotate Trinotate.sqlite \
LOAD_swissprot_blastx swissprot.blastx.outfmt6
% $TRINOTATE_HOME/Trinotate Trinotate.sqlite \
LOAD_swissprot_blastp swissprot.blastp.outfmt6
% $TRINOTATE_HOME/Trinotate Trinotate.sqlite LOAD_pfam TrinotatePFAM.out
% $TRINOTATE_HOME/Trinotate Trinotate.sqlite LOAD_signalp signalp.out
% $TRINOTATE_HOME/Trinotate Trinotate.sqlite report > Trinotate.xls
View the report
% less Trinotate.xls
.
#gene_id transcript_id sprot_Top_BLASTX_hit TrEMBL_Top_BLASTX_hit RNAMMER prot_id prot_coords sprot_Top_BLASTP_hit TrEMBL_Top_BLASTP_hit Pfam SignalP TmHMM eggnog gene_ontology_blast gene_ontology_pfam transcript peptide
TRINITY_DN144_c0_g1 TRINITY_DN144_c0_g1_i1 PUT4_YEAST^PUT4_YEAST^Q:1-198,H:425-490^74.24%ID^E:4e-29^RecName: Full=Proline-specific permease;^Eukaryota; Fungi; Dikarya; Ascomycota; Saccharomycotina; Saccharomycetes; Saccharomycetales; Saccharomycetaceae; Saccharomyces . . . .
. . . . . COG0833^permease GO:0016021^cellular_component^integral component of membrane`GO:0005886^cellular_component^plasma membrane`GO:0015193^molecular_function^L-proline transmembrane transporter activity`GO:0015175^molecular_function^neutral amino acid transmembrane transporter activity`GO:0015812^biological_process^gamma-aminobutyric acid transport`GO:0015804^biological_process^neutral amino acid transport`GO:0035524^biological_process^proline transmembrane transport`GO:0015824^biological_process^proline transport . . .
TRINITY_DN179_c0_g1 TRINITY_DN179_c0_g1_i1 ASNS1_YEAST^ASNS1_YEAST^Q:1-168,H:158-213^82.14%ID^E:5e-30^RecName: Full=Asparagine synthetase [glutamine-hydrolyzing] 1;^Eukaryota; Fungi; Dikarya; Ascomycota; Saccharomycotina; Saccharomycetes; Saccharomycetales; Saccharomycetaceae; Saccharomyces .
. . . . . . . . COG0367^asparagine synthetase GO:0004066^molecular_function^asparagine synthase (glutamine-hydrolyzing) activity`GO:0005524^molecular_function^ATP binding`GO:0006529^biological_process^asparagine biosynthetic process`GO:0006541^biological_process^glutamine metabolic process`GO:0070981^biological_process^L-asparagine biosynthetic process . . .
TRINITY_DN159_c0_g1 TRINITY_DN159_c0_g1_i1 ENO2_CANGA^ENO2_CANGA^Q:2-523,H:128-301^100%ID^E:4e-126^RecName: Full=Enolase 2;^Eukaryota; Fungi; Dikarya; Ascomycota; Saccharomycotina; Saccharomycetes; Saccharomycetales; Saccharomycetaceae; Nakaseomyces; Nakaseomyces/Candida clade . . TRINITY_DN159_c0_g1_i1|m.1 2-523[+] ENO2_CANGA^ENO2_CANGA^Q:1-174,H:128-301^100%ID^E:3e-126^RecName: Full=Enolase 2;^Eukaryota; Fungi; Dikarya; Ascomycota; Saccharomycotina; Saccharomycetes; Saccharomycetales; Saccharomycetaceae; Nakaseomyces; Nakaseomyces/Candida clade . PF00113.17^Enolase_C^Enolase, C-terminal TIM barrel domain^18-174^E:9.2e-79 . . . GO:0005829^cellular_component^cytosol`GO:0000015^cellular_component^phosphopyruvate hydratase complex`GO:0000287^molecular_function^magnesium ion binding`GO:0004634^molecular_function^phosphopyruvate hydratase activity`GO:0006096^biological_process^glycolytic process GO:0000287^molecular_function^magnesium ion binding`GO:0004634^molecular_function^phosphopyruvate hydratase activity`GO:0006096^biological_process^glycolytic process`GO:0000015^cellular_component^phosphopyruvate hydratase complex . .
...
The above file can be very large. It's often useful to load it into a spreadsheet software tools such as MS-Excel. If you have a transcript identifier of interest, you can always just 'grep' to pull out the annotation for that transcript from this report. We'll use TrinotateWeb to interactively explore these data in a web browser below.
Let's use the annotation attributes for the transcripts here as 'names' for the transcripts in the Trinotate database. This will be useful later when using the TrinotateWeb framework.
% $TRINOTATE_HOME/util/annotation_importer/import_transcript_names.pl \
Trinotate.sqlite Trinotate.xls
Nothing exciting to see in running the above command, but know that it's helpful for later on.
Earlier, we generated large sets of tab-delimited files containg lots of data - annotations for transcripts, matrices of expression values, lists of differentially expressed transcripts, etc. We also generated a number of plots in PDF format. These are all useful, but they're not interactive and it's often difficult and cumbersome to extract information of interest during a study. We're developing TrinotateWeb as a web-based interactive system to solve some of these challenges. TrinotateWeb provides heatmaps and various plots of expression data, and includes search functions to quickly access information of interest. Below, we will populate some of the additional information that we need into our Trinotate database, and then run TrinotateWeb and start exploring our data in a web browser.
Once again, verify that you're currently in the Trinotate/ working directory:
% pwd
.
/home/genomics/workshop_materials/transcriptomics/KrumlovTrinityWorkshopJan2018/Trinotate
Now, load in the transcript expression data stored in the matrices we built earlier:
% $TRINOTATE_HOME/util/transcript_expression/import_expression_and_DE_results.pl \
--sqlite Trinotate.sqlite \
--transcript_mode \
--samples_file ../data/samples.txt \
--count_matrix ../Trinity.isoform.counts.matrix \
--fpkm_matrix ../Trinity.isoform.TMM.EXPR.matrix
Import the DE results from our DESeq2_trans/ directory:
% $TRINOTATE_HOME/util/transcript_expression/import_expression_and_DE_results.pl \
--sqlite Trinotate.sqlite \
--transcript_mode \
--samples_file ../data/samples.txt \
--DE_dir ../DESeq2_trans
and Import the clusters of transcripts we extracted earlier based on having similar expression profiles across samples:
% $TRINOTATE_HOME/util/transcript_expression/import_transcript_clusters.pl \
--sqlite Trinotate.sqlite \
--group_name DE_all_vs_all \
--analysis_name diffExpr.P1e-3_C2_clusters_fixed_P_60 \
../DESeq2_trans/diffExpr.P1e-3_C2.matrix.RData.clusters_fixed_P_60/*matrix
And now we'll do the same for our gene-level expression and DE results:
% $TRINOTATE_HOME/util/transcript_expression/import_expression_and_DE_results.pl \
--sqlite Trinotate.sqlite \
--gene_mode \
--samples_file ../data/samples.txt \
--count_matrix ../Trinity.gene.counts.matrix \
--fpkm_matrix ../Trinity.gene.TMM.EXPR.matrix
% $TRINOTATE_HOME/util/transcript_expression/import_expression_and_DE_results.pl \
--sqlite Trinotate.sqlite \
--gene_mode \
--samples_file ../data/samples.txt \
--DE_dir ../DESeq2_gene
Note, in the above gene-loading commands, the term 'component' is used. 'Component' is just another word for 'gene' in the realm of Trinity.
At this point, the Trinotate database should be fully populated and ready to be used by TrinotateWeb.
TrinotateWeb is web-based software and runs locally on the same hardware we've been running all our computes (as opposed to your typical websites that you visit regularly, such as facebook). Launch the mini webserver that drives the TrinotateWeb software like so:
% $TRINOTATE_HOME/run_TrinotateWebserver.pl 8686
Now, visit the following URL in Google Chrome: http://localhost:8686/cgi-bin/index.cgi
You should see a web form like so:

In the text box, put the path to your Trinotate.sqlite database, as shown above ("/home/genomics/workshop_materials/transcriptomics/KrumlovTrinityWorkshopJan2018/Trinotate/Trinotate.sqlite"). Click 'Submit'.
You should now have TrinotateWeb running and serving the content in your Trinotate database:

Take some time to click the various tabs and explore what's available.
eg. Under 'Annotation Keyword Search', search for 'transporter'
eg. Under 'Differential Expression', examine your earlier-defined transcript clusters. Also, launch MA or Volcano plots to explore the DE data.
We will explore TrinotateWeb functionality together as a group.
If you've gotten this far, hurray!!! Congratulations!!! You've now experienced the full tour of Trinity and TrinotateWeb. Visit our web documentation at http://trinityrnaseq.github.io, and join our Google group to become part of the ever-growing Trinity user community.