-
Notifications
You must be signed in to change notification settings - Fork 16
/
16-Faceting.Rmd
232 lines (164 loc) · 7.23 KB
/
16-Faceting.Rmd
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
# Faceting
**Learning objectives:**
- Facet wrap
- Facet grid
- Controlling scales and space
- Missing faceting variables
- Grouping vs. faceting
- Continuous variables
## What is faceting?
```{r message=FALSE, warning=FALSE, paged.print=FALSE}
library(tidyverse)
mpg2 <- subset(mpg, cyl != 5 & drv %in% c("4", "f") & class != "2seater")
```
Faceting breaks down a dataset into multiple plots that show different subsets of the data, often plotting the same variables in each plot.
There are three types of faceting functions:
- `facet_null()`: a single plot, the default
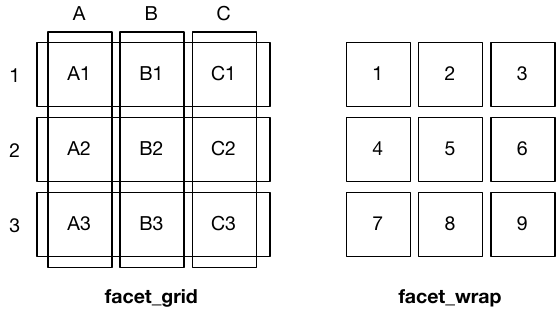
- `facet_wrap()`: “wraps” a 1d ribbon of panels into 2d
- `facet_grid()`: produces a 2d grid of panels defined by variables which form the rows and columns
## Facet wrap
Useful if you have a single variable with many levels and want to arrange the plots in a spatially efficient way. For example, if you have multiple individuals within a study.
Useful arguments:
- `ncol` and `nrow` control how many columns or rows, respectively. Only one of these needs to be set.
- `as.table` controls whether the facets are laid out like a table (`TRUE`, the default), with highest values at the bottom-right, or a plot (`FALSE`), with the highest values at the top-right.
- `dir` controls the direction of wrap: horizontal (`"h"`) or vertical (`"v"`).
```{r}
base <- ggplot(mpg2, aes(displ, hwy)) +
geom_blank() +
xlab(NULL) +
ylab(NULL)
mpg2%>%count(class)
base + facet_wrap(~class, ncol = 3) + labs(title = "ncol = 3")
base + facet_wrap(~class, ncol = 3, as.table = FALSE) + labs(title = "ncol = 3, as.table = FALSE")
base + facet_wrap(~class, nrow = 3) + labs(title = "nrow = 3")
base + facet_wrap(~class, nrow = 3, dir = "v") + labs (title = "nrow = 3, dir = \"v\"")
```
## Facet grid
`facet_grid()` uses a formula (`y ~ x`) to lay out plots in a 2-dimensional grid. This can be useful to compare across combinations of variables.
```{r}
base + facet_grid(. ~ cyl) + labs(title = ". ~ cyl")
base + facet_grid(drv ~ .) + labs(title = "drv ~ .")
base + facet_grid(drv ~ cyl) + labs(title = "drv ~ cyl")
```
Use multiple variables in the rows and columns by "adding" them: `a + b ~ c + d`
Variables specified on rows and columns will be crossed.
```{r}
base + facet_grid(drv ~ cyl + class) + labs(title = "drv ~ cyl + class")
```
## Controlling scales
The `scales` parameter can be used to control whether the position scales are the same in all panels (fixed) or allowed to vary between panels (free).
- `scales = "fixed"`: x and y scales are fixed across all panels.
- `scales = "free_x"`: the x scale is free, and the y scale is fixed.
- `scales = "free_y"`: the y scale is free, and the x scale is fixed.
- `scales = "free"`: x and y scales vary across panels.
```{r}
p <- ggplot(mpg2, aes(cty, hwy)) +
geom_abline() + # I think this defaults to a default of intercept = 0 and slope = 1?
geom_jitter(width = 0.1, height = 0.1)
p + facet_wrap(~cyl) + labs(title = "default (fixed scales)")
p + facet_wrap(~cyl, scales = "free") + labs(title = "free scales")
```
Free scales can be especially useful when comparing multiple time series measured on different scales.
```{r}
economics_long%>%count(date)
ggplot(economics_long, aes(date, value)) +
geom_line() +
facet_wrap(~variable, scales = "free_y", ncol = 1)
```
## Controlling space
`facet_grid()` has an additional parameter: `space`. This takes the same values as `scales`, but when space is “free”, each column (or row) will have width (or height) proportional to the range of the scale for that column (or row).
This is most useful for categorical scales, where we can assign space proportionally based on the number of levels in each facet.
```{r}
mpg2$model <- reorder(mpg2$model, mpg2$cty)
mpg2$manufacturer <- reorder(mpg2$manufacturer, -mpg2$cty)
ggplot(mpg2, aes(cty, model)) +
geom_point() +
facet_grid(manufacturer ~ .) +
theme(strip.text.y = element_text(angle = 0)) +
labs(title = "fixed space")
ggplot(mpg2, aes(cty, model)) +
geom_point() +
facet_grid(manufacturer ~ ., space = "free") +
theme(strip.text.y = element_text(angle = 0)) +
labs(title = "free space only")
ggplot(mpg2, aes(cty, model)) +
geom_point() +
facet_grid(manufacturer ~ ., scales = "free") +
theme(strip.text.y = element_text(angle = 0)) +
labs(title = "free scales only")
ggplot(mpg2, aes(cty, model)) +
geom_point() +
facet_grid(manufacturer ~ ., scales = "free", space = "free") +
theme(strip.text.y = element_text(angle = 0)) +
labs(title = "free scales and space")
```
## Missing faceting variables
When you add a map layer that doesn't contain a variable, ggplot will display the map in every facet: missing faceting variables are treated like they have all values.
This is useful when you want to add annotations that make it easier to compare among facets.
```{r}
df1 <- data.frame(x = 1:3, y = 1:3, gender = c("f", "f", "m"))
df2 <- data.frame(x = 2, y = 2)
ggplot(df1, aes(x, y)) +
geom_point(data = df2, colour = "red", size = 2) +
geom_point() +
facet_wrap(~gender)
```
## Grouping vs. faceting
With faceting, each group is quite far apart in its own panel, and there is no overlap between the groups. **This is good if the groups overlap a lot, but it does make small differences harder to see.**
When using aesthetics to differentiate groups, the groups are close together and may overlap, but **small differences are easier to see**.
```{r}
df <- data.frame(
x = rnorm(120, c(0, 2, 4)),
y = rnorm(120, c(1, 2, 1)),
z = letters[1:3]
)
ggplot(df, aes(x, y)) +
geom_point(aes(colour = z))
```
```{r}
ggplot(df, aes(x, y)) +
geom_point(aes(color=z)) +
facet_wrap(~z)
```
You can help the viewer make comparisons across facets with some thoughtful annotation.
In this example, we show the mean of every group in each panel.
```{r}
df_sum <- df %>%
group_by(z) %>%
summarise(x = mean(x), y = mean(y)) %>%
rename(z2 = z)
ggplot(df, aes(x, y)) +
geom_point() +
geom_point(data = df_sum, aes(colour = z2), size = 4) +
facet_wrap(~z)
```
Alternatively, we could put all data points in each panel, and only colour the focal data.
```{r}
df2 <- dplyr::select(df, -z)
ggplot(df, aes(x, y)) +
geom_point(data = df2, colour = "grey70") +
geom_point(aes(colour = z)) +
facet_wrap(~z)
```
## Continuous variables
To facet continuous variables, you must first discretise them. ggplot2 provides three helper functions to do so:
- Divide the data into `n` bins each of the same length: `cut_interval(x, n)`
- Divide the data into bins of width `width`: `cut_width(x, width)`.
- Divide the data into `n` bins each containing (approximately) the same number of points: `cut_number(x, n = 10)`
Because the faceting formula does not evaluate functions, you must first create a new variable containing the discretised data.
```{r}
# Bins of width 1
mpg2$disp_w <- cut_width(mpg2$displ, 1)
# Six bins of equal length
mpg2$disp_i <- cut_interval(mpg2$displ, 6)
# Six bins containing equal numbers of points
mpg2$disp_n <- cut_number(mpg2$displ, 6)
plot <- ggplot(mpg2, aes(cty, hwy)) +
geom_point() +
labs(x = NULL, y = NULL)
plot + facet_wrap(~disp_w, nrow = 1)
```
## Meeting Videos
### Cohort 1
`r knitr::include_url("https://www.youtube.com/embed/gKVGjht4N20")`