diff --git a/.github/workflows/deploy.yml b/.github/workflows/deploy.yml
index f6f600ae..5449904d 100644
--- a/.github/workflows/deploy.yml
+++ b/.github/workflows/deploy.yml
@@ -79,19 +79,19 @@ jobs:
runs-on: ${{ matrix.os }}
strategy:
matrix:
- python-version: ["3.8", "3.9", "3.10"]
+ python-version: ["3.9", "3.10", "3.11"]
os: [ubuntu-latest]
include:
- os: macos-11
- python-version: "3.10"
+ python-version: "3.11"
- os: macos-latest
- python-version: "3.10"
+ python-version: "3.11"
- os: windows-2019
- python-version: "3.10"
+ python-version: "3.11"
- os: windows-latest
- python-version: "3.10"
+ python-version: "3.11"
- os: ubuntu-20.04
- python-version: "3.10"
+ python-version: "3.11"
steps:
- name: Checkout source
diff --git a/.github/workflows/test.yml b/.github/workflows/test.yml
index 30ddb2b0..d1992835 100644
--- a/.github/workflows/test.yml
+++ b/.github/workflows/test.yml
@@ -50,19 +50,19 @@ jobs:
runs-on: ${{ matrix.os }}
strategy:
matrix:
- python-version: ["3.8", "3.9", "3.10"]
+ python-version: ["3.9", "3.10", "3.11"]
os: [ubuntu-latest]
include:
- os: macos-11
- python-version: "3.10"
+ python-version: "3.11"
- os: macos-latest
- python-version: "3.10"
+ python-version: "3.11"
- os: windows-2019
- python-version: "3.10"
+ python-version: "3.11"
- os: windows-latest
- python-version: "3.10"
+ python-version: "3.11"
- os: ubuntu-20.04
- python-version: "3.10"
+ python-version: "3.11"
steps:
- uses: actions/checkout@v3
@@ -80,23 +80,16 @@ jobs:
cache: "pip"
cache-dependency-path: "pyproject.toml"
- - uses: conda-incubator/setup-miniconda@v2
- with:
- auto-update-conda: true
- python-version: ${{ matrix.python-version }}
-
# these libraries enable testing on Qt on linux
- uses: tlambert03/setup-qt-libs@v1
- # note: if you need dependencies from conda, considering using
- # setup-miniconda: https://github.com/conda-incubator/setup-miniconda
- # and
- # tox-conda: https://github.com/tox-dev/tox-conda
- name: Install dependencies
- run: python -m pip install "tox<4" tox-gh-actions tox-conda
+ run: python -m pip install tox tox-gh-actions
- name: Test with tox
- run: tox
+ run: tox run
+ env:
+ OS: ${{ matrix.os }}
- name: Coverage
uses: codecov/codecov-action@v3
diff --git a/.gitignore b/.gitignore
index f9c3d531..cddb80a1 100644
--- a/.gitignore
+++ b/.gitignore
@@ -1,4 +1,5 @@
__pycache__
+_version.py
.cache
.coverage
.coverage.*
@@ -54,8 +55,7 @@ instance
lib
lib64
local_settings.py
-models/MDCK_*
-models/test_config.json
+models
nosetests.xml
notebooks
parts
@@ -63,5 +63,5 @@ pip-delete-this-directory.txt
pip-log.txt
sdist
target
+user_config.json
var
-_version.py
diff --git a/.napari-hub/DESCRIPTION.md b/.napari-hub/DESCRIPTION.md
index d12f6534..067dc216 100644
--- a/.napari-hub/DESCRIPTION.md
+++ b/.napari-hub/DESCRIPTION.md
@@ -19,7 +19,7 @@ linkages.
We developed `btrack` for cell tracking in time-lapse microscopy data.
-
+
+## Installation
-## associated plugins
+To install the `napari` plugin associated with `btrack` run the command.
+
+```sh
+pip install btrack[napari]
+```
+
+## Example data
+
+You can try out the btrack plugin using sample data:
+
+```sh
+python btrack/napari/examples/show_btrack_widget.py
+```
+
+which will launch `napari` and the `btrack` widget, along with some sample data.
+
+
+## Setting parameters
+
+There are detailed tips and instructions on parameter settings over at the [documentation](https://btrack.readthedocs.io/en/latest/user_guide/index.html).
+
+
+## Associated plugins
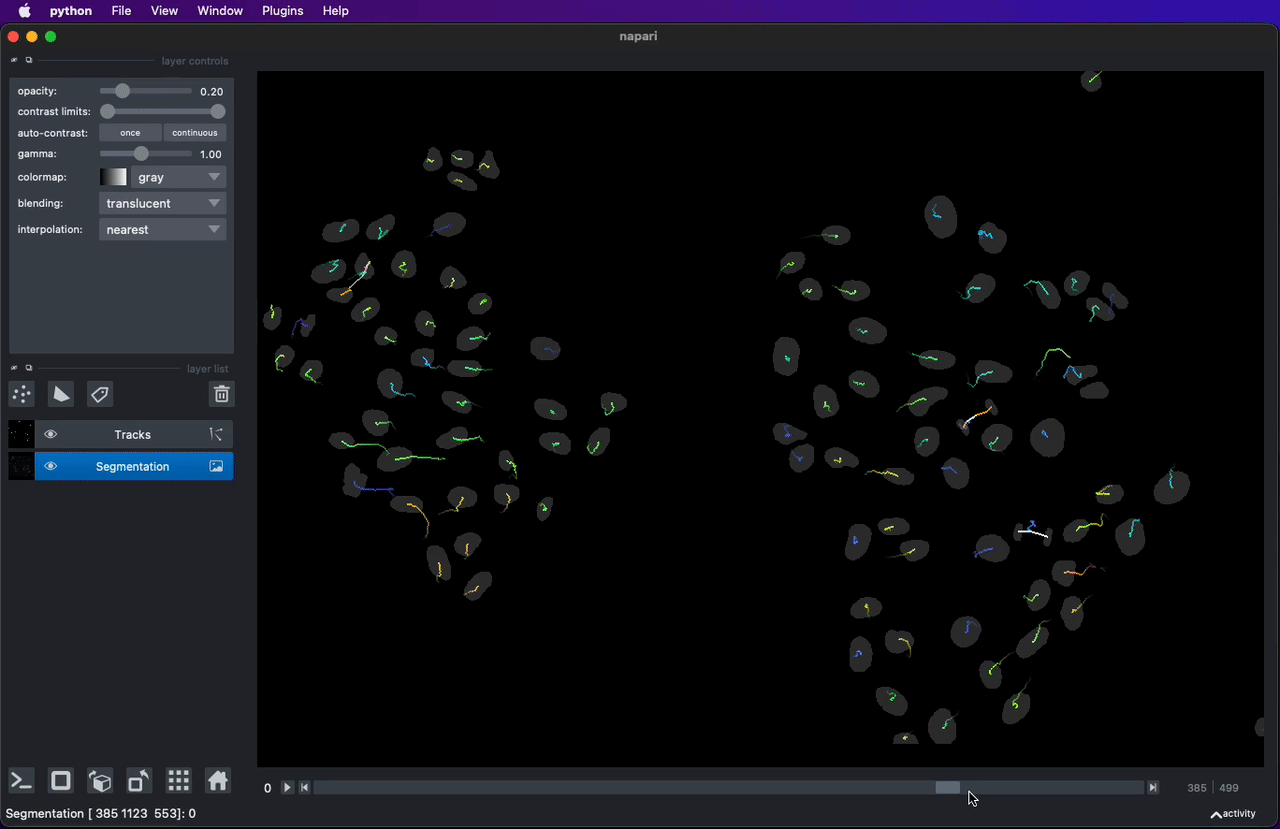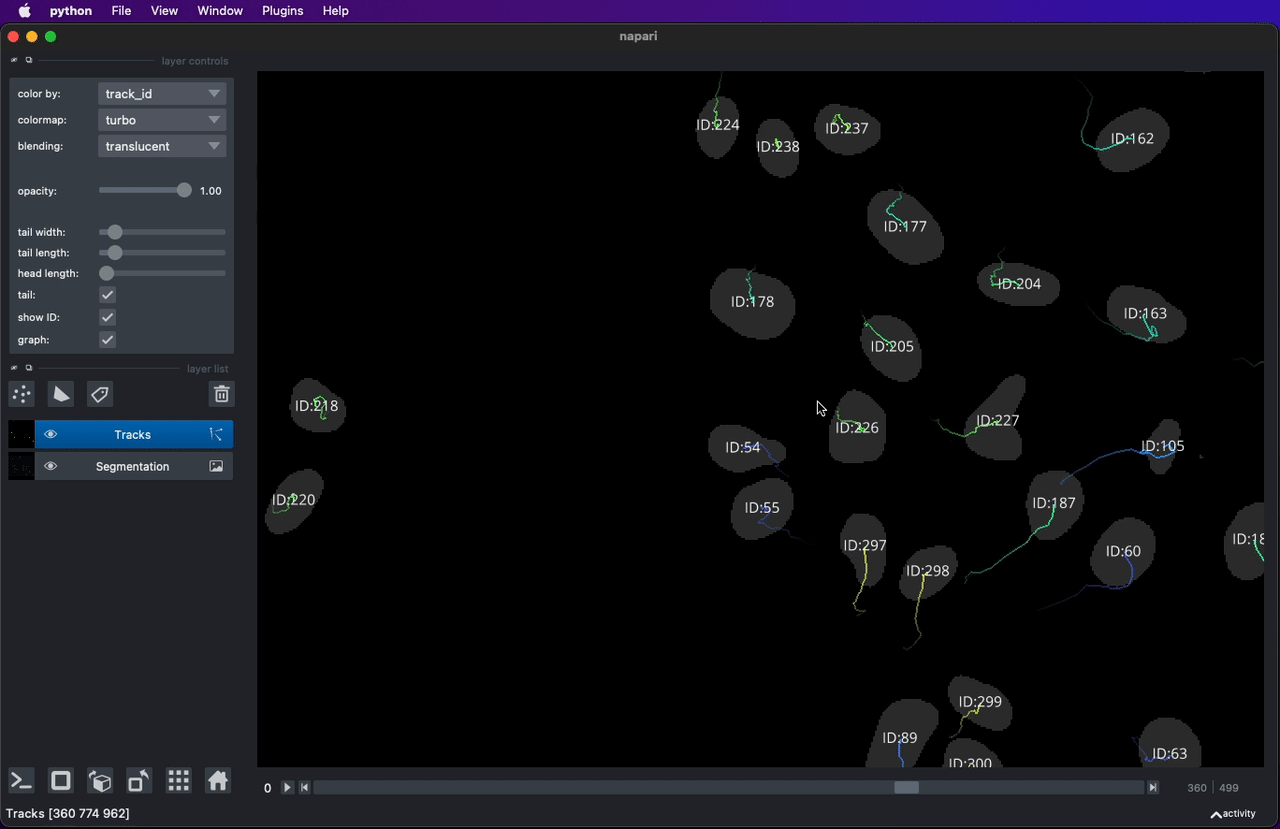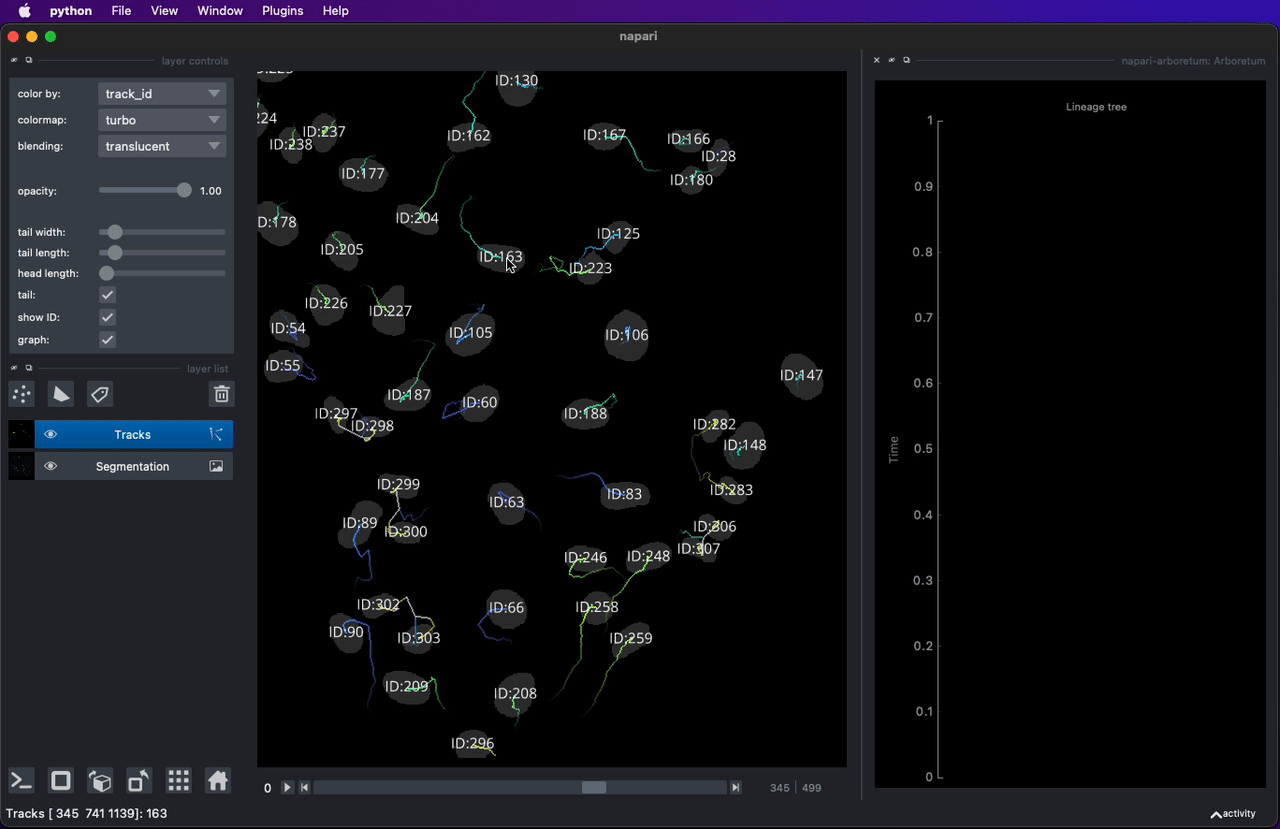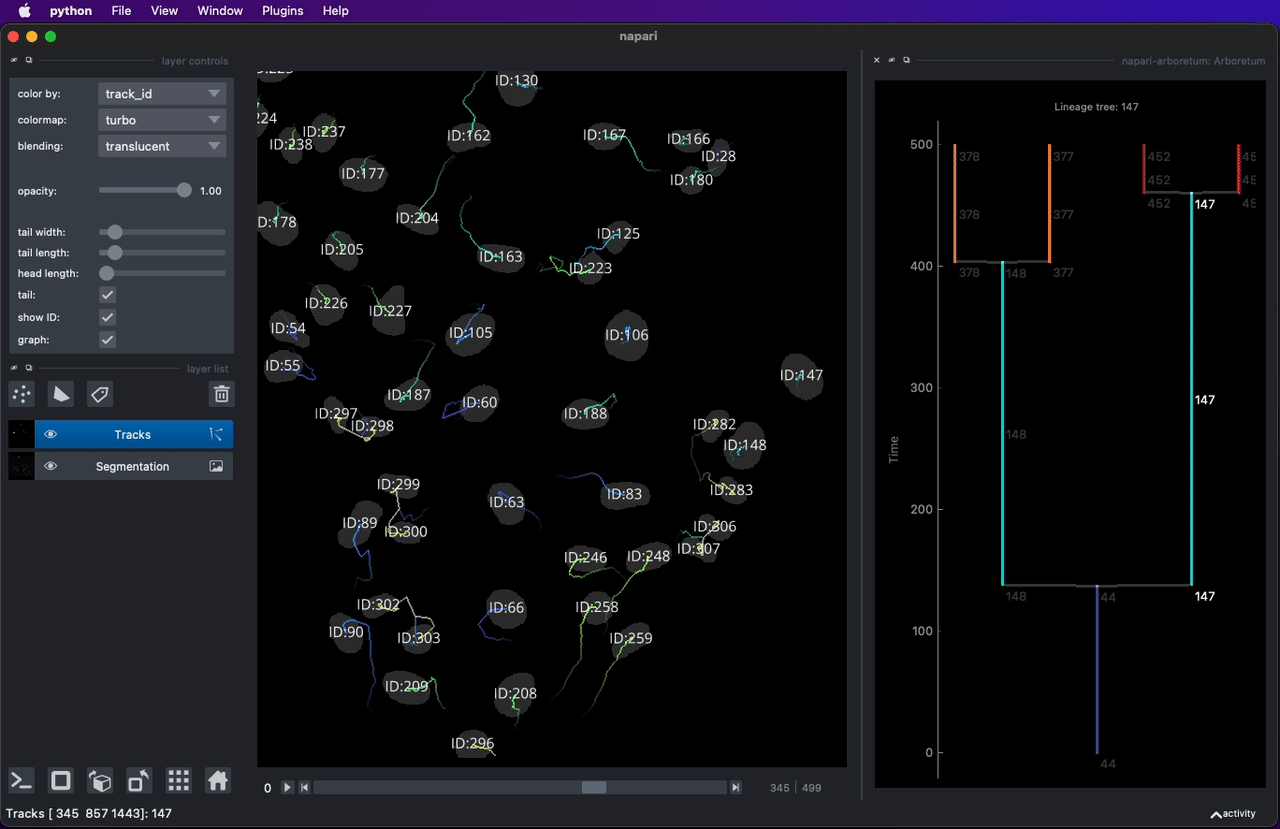
* [napari-arboretum](https://www.napari-hub.org/plugins/napari-arboretum) - Napari plugin to enable track graph and lineage tree visualization.
diff --git a/.pre-commit-config.yaml b/.pre-commit-config.yaml
index 757dcee2..a8de317e 100644
--- a/.pre-commit-config.yaml
+++ b/.pre-commit-config.yaml
@@ -1,10 +1,10 @@
repos:
- repo: https://github.com/charliermarsh/ruff-pre-commit
- rev: v0.0.262
+ rev: v0.0.278
hooks:
- id: ruff
- repo: https://github.com/Lucas-C/pre-commit-hooks
- rev: v1.5.1
+ rev: v1.5.3
hooks:
- id: remove-tabs
exclude: Makefile|docs/Makefile|\.bat$
@@ -19,19 +19,18 @@ repos:
- id: end-of-file-fixer
- id: mixed-line-ending
args: [--fix=lf]
- - id: requirements-txt-fixer
- id: trailing-whitespace
args: [--markdown-linebreak-ext=md]
- repo: https://github.com/psf/black
- rev: 23.3.0
+ rev: 23.7.0
hooks:
- id: black
- repo: https://github.com/pappasam/toml-sort
- rev: v0.23.0
+ rev: v0.23.1
hooks:
- id: toml-sort-fix
- repo: https://github.com/pre-commit/mirrors-clang-format
- rev: v15.0.7
+ rev: v16.0.6
hooks:
- id: clang-format
types_or: [c++, c, cuda]
diff --git a/.readthedocs.yaml b/.readthedocs.yaml
index d15aa007..db1289be 100644
--- a/.readthedocs.yaml
+++ b/.readthedocs.yaml
@@ -9,7 +9,7 @@ version: 2
build:
os: ubuntu-20.04
tools:
- python: "3.10"
+ python: "3.11"
# Optionally declare the Python requirements required to build your docs
python:
diff --git a/MANIFEST.in b/MANIFEST.in
index f3c33ca5..8e90931e 100644
--- a/MANIFEST.in
+++ b/MANIFEST.in
@@ -1 +1,5 @@
graft btrack/libs
+prune docs
+prune tests
+prune models
+prune examples
diff --git a/Makefile b/Makefile
index 1e90e77e..c7afcab0 100644
--- a/Makefile
+++ b/Makefile
@@ -18,15 +18,15 @@ ifeq ($(UNAME), Darwin)
# do something OSX
CXX = clang++ -arch x86_64 -arch arm64
EXT = dylib
- XLD_FLAGS = -arch x86_64 -arch arm64
+ XLDFLAGS =
endif
NVCC = nvcc
-# If your compiler is a bit older you may need to change -std=c++17 to -std=c++0x
+# If your compiler is a bit older you may need to change -std=c++11 to -std=c++0x
#-I/usr/include/python2.7 -L/usr/lib/python2.7 # -O3
LLDBFLAGS =
-CXXFLAGS = -c -std=c++17 -m64 -fPIC -I"./btrack/include" \
+CXXFLAGS = -c -std=c++11 -m64 -fPIC -I"./btrack/include" \
-DDEBUG=false -DBUILD_SHARED_LIB
OPTFLAGS = -O3
LDFLAGS = -shared $(XLDFLAGS)
diff --git a/README.md b/README.md
index d972a8c1..9b89613d 100644
--- a/README.md
+++ b/README.md
@@ -1,5 +1,5 @@
[](https://pypi.org/project/btrack)
-[](https://pepy.tech/project/btrack)
+[](https://pepy.tech/project/btrack)
[](https://github.com/psf/black)
[](https://github.com/quantumjot/btrack/actions/workflows/test.yml)
[](https://github.com/pre-commit/pre-commit)
@@ -32,22 +32,12 @@ Note that `btrack<=0.5.0` was built against earlier version of
[Eigen](https://eigen.tuxfamily.org) which used `C++=11`, as of `btrack==0.5.1`
it is now built against `C++=17`.
-#### Installing the latest stable version
+### Installing the latest stable version
```sh
pip install btrack
```
-## Installing on M1 Mac/Apple Silicon/osx-arm64
-
-Best done with [conda](https://github.com/conda-forge/miniforge)
-
-```sh
-conda env create -f environment.yml
-conda activate btrack
-pip install btrack
-```
-
## Usage examples
Visit [btrack documentation](https://btrack.readthedocs.io) to learn how to use it and see other examples.
diff --git a/btrack/btypes.py b/btrack/btypes.py
index 6921ff3e..b1c3563b 100644
--- a/btrack/btypes.py
+++ b/btrack/btypes.py
@@ -17,9 +17,10 @@
import ctypes
from collections import OrderedDict
-from typing import Any, Dict, List, NamedTuple, Optional, Tuple
+from typing import Any, ClassVar, NamedTuple, Optional
import numpy as np
+from numpy import typing as npt
from . import constants
@@ -27,9 +28,9 @@
class ImagingVolume(NamedTuple):
- x: Tuple[float, float]
- y: Tuple[float, float]
- z: Optional[Tuple[float, float]] = None
+ x: tuple[float, float]
+ y: tuple[float, float]
+ z: Optional[tuple[float, float]] = None
@property
def ndim(self) -> int:
@@ -75,7 +76,7 @@ class PyTrackObject(ctypes.Structure):
Attributes
----------
- properties : Dict[str, Union[int, float]]
+ properties : dict[str, Union[int, float]]
Dictionary of properties associated with this object.
state : constants.States
A state label for the object. See `constants.States`
@@ -86,7 +87,7 @@ class PyTrackObject(ctypes.Structure):
"""
- _fields_ = [
+ _fields_: ClassVar[list] = [
("ID", ctypes.c_long),
("x", ctypes.c_double),
("y", ctypes.c_double),
@@ -108,13 +109,11 @@ def __init__(self):
self._properties = {}
@property
- def properties(self) -> Dict[str, Any]:
- if self.dummy:
- return {}
- return self._properties
+ def properties(self) -> dict[str, Any]:
+ return {} if self.dummy else self._properties
@properties.setter
- def properties(self, properties: Dict[str, Any]):
+ def properties(self, properties: dict[str, Any]):
"""Set the object properties."""
self._properties.update(properties)
@@ -122,17 +121,15 @@ def properties(self, properties: Dict[str, Any]):
def state(self) -> constants.States:
return constants.States(self.label)
- def set_features(self, keys: List[str]) -> None:
+ def set_features(self, keys: list[str]) -> None:
"""Set features to be used by the tracking update."""
if not keys:
self.n_features = 0
return
- if not all(k in self.properties for k in keys):
- missing_features = list(
- set(keys).difference(set(self.properties.keys()))
- )
+ if any(k not in self.properties for k in keys):
+ missing_features = list(set(keys).difference(set(self.properties.keys())))
raise KeyError(f"Feature(s) missing: {missing_features}.")
# store a reference to the numpy array so that Python maintains
@@ -146,18 +143,18 @@ def set_features(self, keys: List[str]) -> None:
self.features = np.ctypeslib.as_ctypes(self._features)
self.n_features = len(self._features)
- def to_dict(self) -> Dict[str, Any]:
+ def to_dict(self) -> dict[str, Any]:
"""Return a dictionary of the fields and their values."""
node = {
k: getattr(self, k)
for k, _ in PyTrackObject._fields_
if k not in ("features", "n_features")
}
- node.update(self.properties)
+ node |= self.properties
return node
@staticmethod
- def from_dict(properties: Dict[str, Any]) -> PyTrackObject:
+ def from_dict(properties: dict[str, Any]) -> PyTrackObject:
"""Build an object from a dictionary."""
obj = PyTrackObject()
fields = dict(PyTrackObject._fields_)
@@ -174,9 +171,7 @@ def from_dict(properties: Dict[str, Any]) -> PyTrackObject:
setattr(obj, key, float(new_data))
# we can add any extra details to the properties dictionary
- obj.properties = {
- k: v for k, v in properties.items() if k not in fields.keys()
- }
+ obj.properties = {k: v for k, v in properties.items() if k not in fields}
return obj
def __repr__(self):
@@ -221,7 +216,7 @@ class PyTrackingInfo(ctypes.Structure):
"""
- _fields_ = [
+ _fields_: ClassVar[list] = [
("error", ctypes.c_uint),
("n_tracks", ctypes.c_uint),
("n_active", ctypes.c_uint),
@@ -235,12 +230,11 @@ class PyTrackingInfo(ctypes.Structure):
("complete", ctypes.c_bool),
]
- def to_dict(self) -> Dict[str, Any]:
+ def to_dict(self) -> dict[str, Any]:
"""Return a dictionary of the statistics"""
# TODO(arl): make this more readable by converting seconds, ms
# and interpreting error messages?
- stats = {k: getattr(self, k) for k, typ in PyTrackingInfo._fields_}
- return stats
+ return {k: getattr(self, k) for k, typ in PyTrackingInfo._fields_}
@property
def tracker_active(self) -> bool:
@@ -269,7 +263,7 @@ class PyGraphEdge(ctypes.Structure):
source timestamp, we just assume that the tracker has done it's job.
"""
- _fields_ = [
+ _fields_: ClassVar[list] = [
("source", ctypes.c_long),
("target", ctypes.c_long),
("score", ctypes.c_double),
@@ -278,8 +272,7 @@ class PyGraphEdge(ctypes.Structure):
def to_dict(self) -> dict[str, Any]:
"""Return a dictionary describing the edge."""
- edge = {k: getattr(self, k) for k, _ in PyGraphEdge._fields_}
- return edge
+ return {k: getattr(self, k) for k, _ in PyGraphEdge._fields_}
class Tracklet:
@@ -312,6 +305,8 @@ class Tracklet:
A list specifying which objects are dummy objects inserted by the tracker.
parent : int, list
The identifiers of the parent track(s).
+ generation : int
+ If specified, the generational depth of the tracklet releative to the root.
refs : list[int]
Returns a list of :py:class:`btrack.btypes.PyTrackObject` identifiers
used to build the track. Useful for indexing back into the original
@@ -323,7 +318,7 @@ class Tracklet:
softmax : list[float]
If defined, return the softmax score for the label of each object in the
track.
- properties : Dict[str, np.ndarray]
+ properties : dict[str, npt.NDArray]
Return a dictionary of track properties derived from
:py:class:`btrack.btypes.PyTrackObject` properties.
root : int,
@@ -336,7 +331,7 @@ class Tracklet:
First time stamp of track.
stop : int, float
Last time stamp of track.
- kalman : np.ndarray
+ kalman : npt.NDArray
Return the complete output of the kalman filter for this track. Note,
that this may not have been returned while from the tracker. See
:py:attr:`btrack.BayesianTracker.return_kalman` for more details.
@@ -353,20 +348,20 @@ class Tracklet:
x values.
"""
- def __init__(
+ def __init__( # noqa: PLR0913
self,
ID: int,
- data: List[PyTrackObject],
+ data: list[PyTrackObject],
*,
parent: Optional[int] = None,
- children: Optional[List[int]] = None,
+ children: Optional[list[int]] = None,
fate: constants.Fates = constants.Fates.UNDEFINED,
):
assert all(isinstance(o, PyTrackObject) for o in data)
self.ID = ID
self._data = data
- self._kalman = None
+ self._kalman = np.empty(0)
self.root = None
self.parent = parent
@@ -385,10 +380,10 @@ def _repr_html_(self):
return _pandas_html_repr(self)
@property
- def properties(self) -> Dict[str, np.ndarray]:
+ def properties(self) -> dict:
"""Return the properties of the objects."""
# find the set of keys, then grab the properties
- keys = set()
+ keys: set = set()
for obj in self._data:
keys.update(obj.properties.keys())
@@ -397,11 +392,7 @@ def properties(self) -> Dict[str, np.ndarray]:
# this to fill the properties array with NaN for dummy objects
property_shapes = {
k: next(
- (
- np.asarray(o.properties[k]).shape
- for o in self._data
- if not o.dummy
- ),
+ (np.asarray(o.properties[k]).shape for o in self._data if not o.dummy),
None,
)
for k in keys
@@ -431,7 +422,7 @@ def properties(self) -> Dict[str, np.ndarray]:
return properties
@properties.setter
- def properties(self, properties: Dict[str, np.ndarray]):
+ def properties(self, properties: dict[str, npt.NDArray]):
"""Store properties associated with this Tracklet."""
# TODO(arl): this will need to set the object properties
pass
@@ -489,51 +480,49 @@ def softmax(self) -> list:
@property
def is_root(self) -> bool:
- return (
- self.parent == 0 or self.parent is None or self.parent == self.ID
- )
+ return self.parent == 0 or self.parent is None or self.parent == self.ID
@property
def is_leaf(self) -> bool:
return not self.children
@property
- def kalman(self) -> np.ndarray:
+ def kalman(self) -> npt.NDArray:
return self._kalman
@kalman.setter
- def kalman(self, data: np.ndarray) -> None:
+ def kalman(self, data: npt.NDArray) -> None:
assert isinstance(data, np.ndarray)
self._kalman = data
- def mu(self, index: int) -> np.ndarray:
+ def mu(self, index: int) -> npt.NDArray:
"""Return the Kalman filter mu. Note that we are only returning the mu
for the positions (e.g. 3x1)."""
return self.kalman[index, 1:4].reshape(3, 1)
- def covar(self, index: int) -> np.ndarray:
+ def covar(self, index: int) -> npt.NDArray:
"""Return the Kalman filter covariance matrix. Note that we are
only returning the covariance matrix for the positions (e.g. 3x3)."""
return self.kalman[index, 4:13].reshape(3, 3)
- def predicted(self, index: int) -> np.ndarray:
+ def predicted(self, index: int) -> npt.NDArray:
"""Return the motion model prediction for the given timestep."""
return self.kalman[index, 13:].reshape(3, 1)
def to_dict(
self, properties: list = constants.DEFAULT_EXPORT_PROPERTIES
- ) -> Dict[str, Any]:
+ ) -> dict[str, Any]:
"""Return a dictionary of the tracklet which can be used for JSON
export. This is an ordered dictionary for nicer JSON output.
"""
- trk_tuple = tuple([(p, getattr(self, p)) for p in properties])
+ trk_tuple = tuple((p, getattr(self, p)) for p in properties)
data = OrderedDict(trk_tuple)
- data.update(self.properties)
+ data |= self.properties
return data
def to_array(
self, properties: list = constants.DEFAULT_EXPORT_PROPERTIES
- ) -> np.ndarray:
+ ) -> npt.NDArray:
"""Return a representation of the trackled as a numpy array."""
data = self.to_dict(properties)
tmp_track = []
@@ -544,10 +533,10 @@ def to_array(
np_values = np.reshape(np_values, (len(self), -1))
tmp_track.append(np_values)
- tmp_track = np.concatenate(tmp_track, axis=-1)
- assert tmp_track.shape[0] == len(self)
- assert tmp_track.ndim == constants.Dimensionality.TWO
- return tmp_track.astype(np.float32)
+ tmp_track_arr = np.concatenate(tmp_track, axis=-1)
+ assert tmp_track_arr.shape[0] == len(self)
+ assert tmp_track_arr.ndim == constants.Dimensionality.TWO
+ return tmp_track_arr.astype(np.float32)
def in_frame(self, frame: int) -> bool:
"""Return true or false as to whether the track is in the frame."""
@@ -558,7 +547,7 @@ def trim(self, frame: int, tail: int = 75) -> Tracklet:
d = [o for o in self._data if o.t <= frame and o.t >= frame - tail]
return Tracklet(self.ID, d)
- def LBEP(self) -> Tuple[int]:
+ def LBEP(self) -> tuple[int, list, list, Optional[int], None, int]:
"""Return an LBEP table summarising the track."""
return (
self.ID,
@@ -576,8 +565,7 @@ def _pandas_html_repr(obj):
import pandas as pd
except ImportError:
return (
- "Install pandas for nicer, tabular rendering.
"
- + obj.__repr__()
+ "Install pandas for nicer, tabular rendering.
" + obj.__repr__()
)
obj_as_dict = obj.to_dict()
diff --git a/btrack/config.py b/btrack/config.py
index f6db4f17..c5aa15e3 100644
--- a/btrack/config.py
+++ b/btrack/config.py
@@ -2,7 +2,7 @@
import logging
import os
from pathlib import Path
-from typing import List, Optional
+from typing import ClassVar, Optional
import numpy as np
from pydantic import BaseModel, conlist, validator
@@ -66,6 +66,9 @@ class TrackerConfig(BaseModel):
tracking_updates : list
A list of features to be used for tracking, such as MOTION or VISUAL.
Must have at least one entry.
+ enable_optimisation
+ A flag which, if `False`, will report a warning to the user if they then
+ subsequently run the `BayesianTracker.optimise()` step.
Notes
-----
@@ -84,7 +87,7 @@ class TrackerConfig(BaseModel):
volume: Optional[ImagingVolume] = None
update_method: constants.BayesianUpdates = constants.BayesianUpdates.EXACT
optimizer_options: dict = constants.GLPK_OPTIONS
- features: List[str] = []
+ features: list[str] = []
tracking_updates: conlist(
constants.BayesianUpdateFeatures,
min_items=1,
@@ -92,20 +95,18 @@ class TrackerConfig(BaseModel):
) = [
constants.BayesianUpdateFeatures.MOTION,
]
+ enable_optimisation = True
@validator("volume", pre=True, always=True)
def _parse_volume(cls, v):
- if isinstance(v, tuple):
- return ImagingVolume(*v)
- return v
+ return ImagingVolume(*v) if isinstance(v, tuple) else v
@validator("tracking_updates", pre=True, always=True)
def _parse_tracking_updates(cls, v):
_tracking_updates = v
if all(isinstance(k, str) for k in _tracking_updates):
_tracking_updates = [
- constants.BayesianUpdateFeatures[k.upper()]
- for k in _tracking_updates
+ constants.BayesianUpdateFeatures[k.upper()] for k in _tracking_updates
]
_tracking_updates = list(set(_tracking_updates))
return _tracking_updates
@@ -113,7 +114,7 @@ def _parse_tracking_updates(cls, v):
class Config:
arbitrary_types_allowed = True
validate_assignment = True
- json_encoders = {
+ json_encoders: ClassVar[dict] = {
np.ndarray: lambda x: x.ravel().tolist(),
}
diff --git a/btrack/constants.py b/btrack/constants.py
index cbe954f3..2f948693 100644
--- a/btrack/constants.py
+++ b/btrack/constants.py
@@ -69,7 +69,7 @@ class Fates(enum.Enum):
@enum.unique
-class States(enum.Enum):
+class States(enum.IntEnum):
INTERPHASE = 0
PROMETAPHASE = 1
METAPHASE = 2
@@ -96,3 +96,4 @@ class Dimensionality(enum.IntEnum):
TWO: int = 2
THREE: int = 3
FOUR: int = 4
+ FIVE: int = 5
diff --git a/btrack/core.py b/btrack/core.py
index 177d7c84..a220eebc 100644
--- a/btrack/core.py
+++ b/btrack/core.py
@@ -3,9 +3,10 @@
import logging
import os
import warnings
-from typing import List, Optional, Tuple, Union
+from typing import Optional, Union
import numpy as np
+from numpy import typing as npt
from btrack import _version
@@ -49,7 +50,7 @@ class BayesianTracker:
:py:meth:`btrack.btypes.ImagingVolume` for more details.
frame_range : tuple
The frame range for tracking, essentially the last dimension of volume.
- LBEP : List[List]
+ LBEP : list[List]
Return an LBEP table of the track lineages.
configuration : config.TrackerConfig
Return the current configuration.
@@ -137,12 +138,10 @@ def __init__(
self._config = config.TrackerConfig(verbose=verbose)
# silently set the update method to EXACT
- self._lib.set_update_mode(
- self._engine, self.configuration.update_method.value
- )
+ self._lib.set_update_mode(self._engine, self.configuration.update_method.value)
# default parameters and space for stored objects
- self._objects: List[btypes.PyTrackObject] = []
+ self._objects: list[btypes.PyTrackObject] = []
self._frame_range = [0, 0]
def __enter__(self):
@@ -224,7 +223,7 @@ def _max_search_radius(self, max_search_radius: int):
"""Set the maximum search radius for fast cost updates."""
self._lib.max_search_radius(self._engine, max_search_radius)
- def _update_method(self, method: Union[str, constants.BayesianUpdates]):
+ def _update_method(self, method: constants.BayesianUpdates):
"""Set the method for updates, EXACT, APPROXIMATE, CUDA etc..."""
self._lib.set_update_mode(self._engine, method.value)
@@ -243,12 +242,10 @@ def n_tracks(self) -> int:
@property
def n_dummies(self) -> int:
"""Return the number of dummy objects (negative ID)."""
- return len(
- [d for d in itertools.chain.from_iterable(self.refs) if d < 0]
- )
+ return len([d for d in itertools.chain.from_iterable(self.refs) if d < 0])
@property
- def tracks(self) -> List[btypes.Tracklet]:
+ def tracks(self) -> list[btypes.Tracklet]:
"""Return a sorted list of tracks, default is to sort by increasing
length."""
return [self[i] for i in range(self.n_tracks)]
@@ -273,8 +270,7 @@ def refs(self):
def dummies(self):
"""Return a list of dummy objects."""
return [
- self._lib.get_dummy(self._engine, -(i + 1))
- for i in range(self.n_dummies)
+ self._lib.get_dummy(self._engine, -(i + 1)) for i in range(self.n_dummies)
]
@property
@@ -305,7 +301,7 @@ def LBEP(self):
"""
return utils._lbep_table(self.tracks)
- def _sort(self, tracks: List[btypes.Tracklet]) -> List[btypes.Tracklet]:
+ def _sort(self, tracks: list[btypes.Tracklet]) -> list[btypes.Tracklet]:
"""Return a sorted list of tracks"""
return sorted(tracks, key=lambda t: len(t), reverse=True)
@@ -374,18 +370,16 @@ def _object_model(self, model: models.ObjectModel) -> None:
)
@property
- def frame_range(self) -> Tuple[int, int]:
+ def frame_range(self) -> tuple:
"""Return the frame range."""
return tuple(self.configuration.frame_range)
@property
- def objects(self) -> List[btypes.PyTrackObject]:
+ def objects(self) -> list[btypes.PyTrackObject]:
"""Return the list of objects added through the append method."""
return self._objects
- def append(
- self, objects: Union[List[btypes.PyTrackObject], np.ndarray]
- ) -> None:
+ def append(self, objects: Union[list[btypes.PyTrackObject], npt.NDArray]) -> None:
"""Append a single track object, or list of objects to the stack. Note
that the tracker will automatically order these by frame number, so the
order here does not matter. This means several datasets can be
@@ -393,7 +387,7 @@ def append(
Parameters
----------
- objects : list, np.ndarray
+ objects : list, npt.NDArray
A list of objects to track.
"""
@@ -424,9 +418,7 @@ def _stats(self, info_ptr: ctypes.pointer) -> btypes.PyTrackingInfo:
return info_ptr.contents
def track_interactive(self, *args, **kwargs) -> None:
- logger.warning(
- "`track_interactive` will be deprecated. Use `track` instead."
- )
+ logger.warning("`track_interactive` will be deprecated. Use `track` instead.")
return self.track(*args, **kwargs)
def track(
@@ -434,7 +426,7 @@ def track(
*,
step_size: int = 100,
tracking_updates: Optional[
- List[Union[str, constants.BayesianUpdateFeatures]]
+ list[Union[str, constants.BayesianUpdateFeatures]]
] = None,
) -> None:
"""Run the tracking in an interactive mode.
@@ -462,7 +454,7 @@ def track(
# bitwise OR is equivalent to int sum here
self._lib.set_update_features(
self._engine,
- sum([int(f.value) for f in self.configuration.tracking_updates]),
+ sum(int(f.value) for f in self.configuration.tracking_updates),
)
stats = self.step()
@@ -488,8 +480,7 @@ def track(
f"(in {stats.t_total_time}s)"
)
logger.info(
- f" - Inserted {self.n_dummies} dummy objects to fill "
- "tracking gaps"
+ f" - Inserted {self.n_dummies} dummy objects to fill tracking gaps"
)
def step(self, n_steps: int = 1) -> Optional[btypes.PyTrackingInfo]:
@@ -499,7 +490,7 @@ def step(self, n_steps: int = 1) -> Optional[btypes.PyTrackingInfo]:
return None
return self._stats(self._lib.step(self._engine, n_steps))
- def hypotheses(self) -> List[hypothesis.Hypothesis]:
+ def hypotheses(self) -> list[hypothesis.Hypothesis]:
"""Calculate and return hypotheses using the hypothesis engine."""
if not self.hypothesis_model:
@@ -513,19 +504,13 @@ def hypotheses(self) -> List[hypothesis.Hypothesis]:
)
# now get all of the hypotheses
- h = [
- self._lib.get_hypothesis(self._engine, h)
- for h in range(n_hypotheses)
- ]
- return h
+ return [self._lib.get_hypothesis(self._engine, h) for h in range(n_hypotheses)]
def optimize(self, **kwargs):
"""Proxy for `optimise` for our American friends ;)"""
return self.optimise(**kwargs)
- def optimise(
- self, options: Optional[dict] = None
- ) -> List[hypothesis.Hypothesis]:
+ def optimise(self, options: Optional[dict] = None) -> list[hypothesis.Hypothesis]:
"""Optimize the tracks.
Parameters
@@ -544,19 +529,18 @@ def optimise(
optimiser and then performs track merging, removal of track fragments,
renumbering and assignment of branches.
"""
+ if not self.configuration.enable_optimisation:
+ logger.warning("The `enable_optimisation` flag is set to False")
+
logger.info(f"Loading hypothesis model: {self.hypothesis_model.name}")
- logger.info(
- f"Calculating hypotheses (relax: {self.hypothesis_model.relax})..."
- )
+ logger.info(f"Calculating hypotheses (relax: {self.hypothesis_model.relax})...")
hypotheses = self.hypotheses()
# if we have not been provided with optimizer options, use the default
# from the configuration.
options = (
- options
- if options is not None
- else self.configuration.optimizer_options
+ options if options is not None else self.configuration.optimizer_options
)
# if we don't have any hypotheses return
@@ -673,25 +657,22 @@ def export(
A string that represents how the data has been filtered prior to
tracking, e.g. using the object property `area>100`
"""
- export_delegator(
- filename, self, obj_type=obj_type, filter_by=filter_by
- )
+ export_delegator(filename, self, obj_type=obj_type, filter_by=filter_by)
def to_napari(
self,
replace_nan: bool = True, # noqa: FBT001,FBT002
ndim: Optional[int] = None,
- ) -> Tuple[np.ndarray, dict, dict]:
+ ) -> tuple[npt.NDArray, dict, dict]:
"""Return the data in a format for a napari tracks layer.
See :py:meth:`btrack.utils.tracks_to_napari`."""
+ assert self.configuration.volume is not None
ndim = self.configuration.volume.ndim if ndim is None else ndim
- return utils.tracks_to_napari(
- self.tracks, ndim=ndim, replace_nan=replace_nan
- )
+ return utils.tracks_to_napari(self.tracks, ndim=ndim, replace_nan=replace_nan)
- def candidate_graph_edges(self) -> List[btypes.PyGraphEdge]:
+ def candidate_graph_edges(self) -> list[btypes.PyGraphEdge]:
"""Return the edges from the full candidate graph."""
num_edges = self._lib.num_edges(self._engine)
if num_edges < 1:
@@ -700,7 +681,4 @@ def candidate_graph_edges(self) -> List[btypes.PyGraphEdge]:
"``config.store_candidate_graph`` is set to "
f"{self.configuration.store_candidate_graph}"
)
- return [
- self._lib.get_graph_edge(self._engine, idx)
- for idx in range(num_edges)
- ]
+ return [self._lib.get_graph_edge(self._engine, idx) for idx in range(num_edges)]
diff --git a/btrack/datasets.py b/btrack/datasets.py
index 585ae0c0..2d89763e 100644
--- a/btrack/datasets.py
+++ b/btrack/datasets.py
@@ -1,28 +1,24 @@
import os
-from typing import List
-import numpy as np
import pooch
+from numpy import typing as npt
from skimage.io import imread
-from .btypes import PyTrackObject
-from .io import import_CSV
+from .btypes import PyTrackObject, Tracklet
+from .io import HDF5FileHandler, import_CSV
-BASE_URL = (
- "https://raw.githubusercontent.com/lowe-lab-ucl/btrack-examples/main/"
-)
+BASE_URL = "https://raw.githubusercontent.com/lowe-lab-ucl/btrack-examples/main/"
CACHE_PATH = pooch.os_cache("btrack-examples")
def _remote_registry() -> os.PathLike:
- file_path = pooch.retrieve(
- # URL to one of Pooch's test files
+ # URL to one of Pooch's test files
+ return pooch.retrieve(
path=CACHE_PATH,
- url=BASE_URL + "registry.txt",
- known_hash="673de62c62eeb6f356fb1bff968748566d23936f567201cf61493d031d42d480",
+ url=f"{BASE_URL}registry.txt",
+ known_hash="20d8c44289f421ab52d109e6af2c76610e740230479fe5c46a4e94463c9b5d50",
)
- return file_path
POOCH = pooch.create(
@@ -36,39 +32,42 @@ def _remote_registry() -> os.PathLike:
def cell_config() -> os.PathLike:
"""Return the file path to the example `cell_config`."""
- file_path = POOCH.fetch("examples/cell_config.json")
- return file_path
+ return POOCH.fetch("examples/cell_config.json")
def particle_config() -> os.PathLike:
"""Return the file path to the example `particle_config`."""
- file_path = POOCH.fetch("examples/particle_config.json")
- return file_path
+ return POOCH.fetch("examples/particle_config.json")
def example_segmentation_file() -> os.PathLike:
"""Return the file path to the example U-Net segmentation image file."""
- file_path = POOCH.fetch("examples/segmented.tif")
- return file_path
+ return POOCH.fetch("examples/segmented.tif")
-def example_segmentation() -> np.ndarray:
+def example_segmentation() -> npt.NDArray:
"""Return the U-Net segmentation as a numpy array of dimensions (T, Y, X)."""
file_path = example_segmentation_file()
- segmentation = imread(file_path)
- return segmentation
+ return imread(file_path)
def example_track_objects_file() -> os.PathLike:
"""Return the file path to the example localized and classified objects
stored in a CSV file."""
- file_path = POOCH.fetch("examples/objects.csv")
- return file_path
+ return POOCH.fetch("examples/objects.csv")
-def example_track_objects() -> List[PyTrackObject]:
+def example_track_objects() -> list[PyTrackObject]:
"""Return the example localized and classified objects stored in a CSV file
as a list `PyTrackObject`s to be used by the tracker."""
file_path = example_track_objects_file()
- objects = import_CSV(file_path)
- return objects
+ return import_CSV(file_path)
+
+
+def example_tracks() -> list[Tracklet]:
+ """Return the example example localized and classified objected stored in an
+ HDF5 file as a list of `Tracklet`s."""
+ file_path = POOCH.fetch("examples/tracks.h5")
+ with HDF5FileHandler(file_path, "r", obj_type="obj_type_1") as reader:
+ tracks = reader.tracks
+ return tracks
diff --git a/btrack/include/tracker.h b/btrack/include/tracker.h
index ff2eb23f..db314c3f 100644
--- a/btrack/include/tracker.h
+++ b/btrack/include/tracker.h
@@ -20,7 +20,7 @@
#include
#include
#include
-#include
+// #include
#include
#include
#include
@@ -264,7 +264,7 @@ class BayesianTracker : public UpdateFeatures {
PyTrackInfo statistics;
// member variable to store an output path for debugging
- std::filesystem::path m_debug_filepath;
+ // std::experimental::filesystem::path m_debug_filepath;
};
// utils to write out belief matrix to CSV files
diff --git a/btrack/io/_localization.py b/btrack/io/_localization.py
index cfeaaff8..cb2da3da 100644
--- a/btrack/io/_localization.py
+++ b/btrack/io/_localization.py
@@ -2,13 +2,18 @@
import dataclasses
import logging
+from collections.abc import Generator
from multiprocessing.pool import Pool
-from typing import Callable, Dict, Generator, List, Optional, Tuple, Union
+from typing import Callable, Optional, Union
import numpy as np
import numpy.typing as npt
from skimage.measure import label, regionprops, regionprops_table
-from tqdm import tqdm
+
+try:
+ from napari.utils import progress as tqdm
+except ImportError:
+ from tqdm import tqdm
from btrack import btypes
from btrack.constants import Dimensionality
@@ -25,13 +30,11 @@ def _is_unique(x: npt.NDArray) -> bool:
def _concat_nodes(
- nodes: Dict[str, npt.NDArray], new_nodes: Dict[str, npt.NDArray]
-) -> Dict[str, npt.NDArray]:
+ nodes: dict[str, npt.NDArray], new_nodes: dict[str, npt.NDArray]
+) -> dict[str, npt.NDArray]:
"""Concatentate centroid dictionaries."""
for key, values in new_nodes.items():
- nodes[key] = (
- np.concatenate([nodes[key], values]) if key in nodes else values
- )
+ nodes[key] = np.concatenate([nodes[key], values]) if key in nodes else values
return nodes
@@ -44,21 +47,17 @@ class SegmentationContainer:
def __post_init__(self) -> None:
self._is_generator = isinstance(self.segmentation, Generator)
- self._next = (
- self._next_generator if self._is_generator else self._next_array
- )
+ self._next = self._next_generator if self._is_generator else self._next_array
- def _next_generator(self) -> Tuple[npt.NDArray, Optional[npt.NDArray]]:
+ def _next_generator(self) -> tuple[npt.NDArray, Optional[npt.NDArray]]:
"""__next__ method for a generator input."""
seg = next(self.segmentation)
intens = (
- next(self.intensity_image)
- if self.intensity_image is not None
- else None
+ next(self.intensity_image) if self.intensity_image is not None else None
)
return seg, intens
- def _next_array(self) -> Tuple[npt.NDArray, Optional[npt.NDArray]]:
+ def _next_array(self) -> tuple[npt.NDArray, Optional[npt.NDArray]]:
"""__next__ method for an array-like input."""
if self._iter >= len(self):
raise StopIteration
@@ -74,7 +73,7 @@ def __iter__(self) -> SegmentationContainer:
self._iter = 0
return self
- def __next__(self) -> Tuple[int, npt.NDArray, Optional[npt.NDArray]]:
+ def __next__(self) -> tuple[int, npt.NDArray, Optional[npt.NDArray]]:
seg, intens = self._next()
data = (self._iter, seg, intens)
self._iter += 1
@@ -88,26 +87,23 @@ def __len__(self) -> int:
class NodeProcessor:
"""Processor to extract nodes from a segmentation image."""
- properties: Tuple[str]
+ properties: tuple[str, ...]
centroid_type: str = "centroid"
intensity_image: Optional[npt.NDArray] = None
- scale: Optional[Tuple[float]] = None
+ scale: Optional[tuple[float]] = None
assign_class_ID: bool = False # noqa: N815
- extra_properties: Optional[Tuple[Callable]] = None
+ extra_properties: Optional[tuple[Callable]] = None
@property
- def img_props(self) -> List[str]:
+ def img_props(self) -> tuple[str, ...]:
# need to infer the name of the function provided
- extra_img_props = tuple(
- [str(fn.__name__) for fn in self.extra_properties]
+ return self.properties + (
+ tuple(str(fn.__name__) for fn in self.extra_properties)
if self.extra_properties
- else []
+ else ()
)
- return self.properties + extra_img_props
- def __call__(
- self, data: Tuple[int, npt.NDAarray, Optional[npt.NDArray]]
- ) -> Dict[str, npt.NDArray]:
+ def __call__(self, data: tuple[int, npt.NDArray, Optional[npt.NDArray]]) -> dict:
"""Return the object centroids from a numpy array representing the
image data."""
@@ -119,33 +115,20 @@ def __call__(
if segmentation.ndim not in (Dimensionality.TWO, Dimensionality.THREE):
raise ValueError("Segmentation array must have 3 or 4 dims.")
- labeled = (
- segmentation if _is_unique(segmentation) else label(segmentation)
- )
+ labeled = segmentation if _is_unique(segmentation) else label(segmentation)
props = regionprops(
labeled,
intensity_image=intensity_image,
extra_properties=self.extra_properties,
)
num_nodes = len(props)
- scale = (
- tuple([1.0] * segmentation.ndim)
- if self.scale is None
- else self.scale
- )
+ scale = tuple([1.0] * segmentation.ndim) if self.scale is None else self.scale
if len(scale) != segmentation.ndim:
- raise ValueError(
- f"Scale dimensions do not match segmentation: {scale}."
- )
+ raise ValueError(f"Scale dimensions do not match segmentation: {scale}.")
centroids = list(
- zip(
- *[
- getattr(props[idx], self.centroid_type)
- for idx in range(num_nodes)
- ]
- )
+ zip(*[getattr(props[idx], self.centroid_type) for idx in range(num_nodes)])
)[::-1]
centroid_dims = ["x", "y", "z"][: segmentation.ndim]
@@ -154,9 +137,7 @@ def __call__(
for dim in range(len(centroids))
}
- nodes = {"t": [frame] * num_nodes}
- nodes.update(coords)
-
+ nodes = {"t": [frame] * num_nodes} | coords
for img_prop in self.img_props:
nodes[img_prop] = [
getattr(props[idx], img_prop) for idx in range(num_nodes)
@@ -173,25 +154,25 @@ def __call__(
return nodes
-def segmentation_to_objects(
- segmentation: Union[np.ndarray, Generator],
+def segmentation_to_objects( # noqa: PLR0913
+ segmentation: Union[npt.NDArray, Generator],
*,
- intensity_image: Optional[Union[np.ndarray, Generator]] = None,
- properties: Optional[Tuple[str]] = (),
- extra_properties: Optional[Tuple[Callable]] = None,
- scale: Optional[Tuple[float]] = None,
+ intensity_image: Optional[Union[npt.NDArray, Generator]] = None,
+ properties: tuple[str, ...] = (),
+ extra_properties: Optional[tuple[Callable]] = None,
+ scale: Optional[tuple[float]] = None,
use_weighted_centroid: bool = True,
assign_class_ID: bool = False,
num_workers: int = 1,
-) -> List[btypes.PyTrackObject]:
+) -> list[btypes.PyTrackObject]:
"""Convert segmentation to a set of trackable objects.
Parameters
----------
- segmentation : np.ndarray, dask.array.core.Array or Generator
+ segmentation : npt.NDArray, dask.array.core.Array or Generator
Segmentation can be provided in several different formats. Arrays should
be ordered as T(Z)YX.
- intensity_image : np.ndarray, dask.array.core.Array or Generator, optional
+ intensity_image : npt.NDArray, dask.array.core.Array or Generator, optional
Intensity image with same size as segmentation, to be used to calculate
additional properties. See `skimage.measure.regionprops` for more info.
properties : tuple of str, optional
@@ -274,11 +255,11 @@ def segmentation_to_objects(
# we need to remove 'label' since this is a protected keyword for btrack
# objects
- if isinstance(properties, tuple) and "label" in properties:
+ if "label" in properties:
logger.warning("Cannot use `scikit-image` `label` as a property.")
- properties = set(properties)
- properties.remove("label")
- properties = tuple(properties)
+ properties_set = set(properties)
+ properties_set.remove("label")
+ properties = tuple(properties_set)
processor = NodeProcessor(
properties=properties,
@@ -297,14 +278,18 @@ def segmentation_to_objects(
num_workers = 1
if num_workers <= 1:
- for data in tqdm(container, total=len(container)):
+ for data in tqdm(container, total=len(container), position=0):
_nodes = processor(data)
nodes = _concat_nodes(nodes, _nodes)
else:
logger.info(f"Processing using {num_workers} workers.")
with Pool(processes=num_workers) as pool:
result = list(
- tqdm(pool.imap(processor, container), total=len(container))
+ tqdm(
+ pool.imap(processor, container),
+ total=len(container),
+ position=0,
+ )
)
for _nodes in result:
diff --git a/btrack/io/exporters.py b/btrack/io/exporters.py
index 5dbfa16c..806de6eb 100644
--- a/btrack/io/exporters.py
+++ b/btrack/io/exporters.py
@@ -3,6 +3,7 @@
import csv
import logging
import os
+from pathlib import Path
from typing import TYPE_CHECKING, Optional
import numpy as np
@@ -112,13 +113,16 @@ def export_LBEP(filename: os.PathLike, tracks: list):
def _export_HDF(
- filename: os.PathLike, tracker, obj_type=None, filter_by: str = None
+ filename: os.PathLike,
+ tracker,
+ obj_type: Optional[str] = None,
+ filter_by: Optional[str] = None,
):
"""Export to HDF."""
filename_noext, ext = os.path.splitext(filename)
if ext != ".h5":
- filename = filename_noext + ".h5"
+ filename = Path(f"{filename_noext}.h5")
logger.warning(f"Changing HDF filename to {filename}")
with HDF5FileHandler(filename, read_write="a", obj_type=obj_type) as hdf:
diff --git a/btrack/io/hdf.py b/btrack/io/hdf.py
index 52697cbf..20cdd6ec 100644
--- a/btrack/io/hdf.py
+++ b/btrack/io/hdf.py
@@ -4,11 +4,13 @@
import logging
import os
import re
+from ast import literal_eval
from functools import wraps
-from typing import TYPE_CHECKING, Any, Dict, List, Optional, Union
+from typing import TYPE_CHECKING, Any, Optional, Union
import h5py
import numpy as np
+from numpy import typing as npt
# import core
from btrack import _version, btypes, constants, utils
@@ -36,9 +38,7 @@ def wrapped_handler_property(*args, **kwargs):
self = args[0]
assert isinstance(self, HDF5FileHandler)
if property not in self._hdf:
- logger.error(
- f"{property.capitalize()} not found in {self.filename}"
- )
+ logger.error(f"{property.capitalize()} not found in {self.filename}")
return None
return fn(*args, **kwargs)
@@ -63,15 +63,15 @@ class HDF5FileHandler:
Attributes
----------
- segmentation : np.ndarray
+ segmentation : npt.NDArray
A numpy array representing the segmentation data. TZYX
objects : list [PyTrackObject]
A list of PyTrackObjects localised from the segmentation data.
- filtered_objects : np.ndarray
+ filtered_objects : npt.NDArray
Similar to objects, but filtered by property.
tracks : list [Tracklet]
A list of Tracklet objects.
- lbep : np.ndarray
+ lbep : npt.NDArray
The LBEP table representing the track graph.
Notes
@@ -140,7 +140,7 @@ def __init__(
self._states = list(constants.States)
@property
- def object_types(self) -> List[str]:
+ def object_types(self) -> list[str]:
return list(self._hdf["objects"].keys())
def __enter__(self):
@@ -167,17 +167,17 @@ def object_type(self, obj_type: str) -> None:
@property # type: ignore
@h5check_property_exists("segmentation")
- def segmentation(self) -> np.ndarray:
+ def segmentation(self) -> npt.NDArray:
segmentation = self._hdf["segmentation"]["images"][:].astype(np.uint16)
logger.info(f"Loading segmentation {segmentation.shape}")
return segmentation
- def write_segmentation(self, segmentation: np.ndarray) -> None:
+ def write_segmentation(self, segmentation: npt.NDArray) -> None:
"""Write out the segmentation to an HDF file.
Parameters
----------
- segmentation : np.ndarray
+ segmentation : npt.NDArray
A numpy array representing the segmentation data. T(Z)YX, uint16
"""
# write the segmentation out
@@ -191,7 +191,7 @@ def write_segmentation(self, segmentation: np.ndarray) -> None:
)
@property
- def objects(self) -> List[btypes.PyTrackObject]:
+ def objects(self) -> list[btypes.PyTrackObject]:
"""Return the objects in the file."""
return self.filtered_objects()
@@ -201,8 +201,8 @@ def filtered_objects(
f_expr: Optional[str] = None,
*,
lazy_load_properties: bool = True,
- exclude_properties: Optional[List[str]] = None,
- ) -> List[btypes.PyTrackObject]:
+ exclude_properties: Optional[list[str]] = None,
+ ) -> list[btypes.PyTrackObject]:
"""A filtered list of objects based on metadata.
Parameters
@@ -244,9 +244,7 @@ def filtered_objects(
properties = {}
if "properties" in grp:
p_keys = list(
- set(grp["properties"].keys()).difference(
- set(exclude_properties)
- )
+ set(grp["properties"].keys()).difference(set(exclude_properties))
)
properties = {k: grp["properties"][k][:] for k in p_keys}
assert all(len(p) == len(txyz) for p in properties.values())
@@ -263,14 +261,28 @@ def filtered_objects(
f_eval = f"x{m['op']}{m['cmp']}" # e.g. x > 10
+ data = None
+
if m["name"] in properties:
data = properties[m["name"]]
- filtered_idx = [i for i, x in enumerate(data) if eval(f_eval)]
+ elif m["name"] in grp:
+ logger.warning(
+ f"While trying to filter objects by `{f_expr}` encountered "
+ "a legacy HDF file."
+ )
+ logger.warning(
+ "Properties do not persist to objects. Use `hdf.tree()` to "
+ "inspect the file structure."
+ )
+ data = grp[m["name"]]
else:
raise ValueError(f"Cannot filter objects by {f_expr}")
+ filtered_idx = [i for i, x in enumerate(data) if literal_eval(f_eval)]
+
else:
- filtered_idx = range(txyz.shape[0]) # default filtering uses all
+ # default filtering uses all
+ filtered_idx = list(range(txyz.shape[0]))
# sanity check that coordinates matches labels
assert txyz.shape[0] == labels.shape[0]
@@ -293,12 +305,12 @@ def filtered_objects(
# add the filtered properties
for key, props in properties.items():
- objects_dict.update({key: props[filtered_idx]})
+ objects_dict[key] = props[filtered_idx]
return objects_from_dict(objects_dict)
def write_objects(
- self, data: Union[List[btypes.PyTrackObject], BayesianTracker]
+ self, data: Union[list[btypes.PyTrackObject], BayesianTracker]
) -> None:
"""Write objects to HDF file.
@@ -324,7 +336,7 @@ def write_objects(
if "objects" not in self._hdf:
self._hdf.create_group("objects")
grp = self._hdf["objects"].create_group(self.object_type)
- props = {k: [] for k in objects[0].properties}
+ props: dict = {k: [] for k in objects[0].properties}
n_objects = len(objects)
n_frames = np.max([o.t for o in objects]) + 1
@@ -358,7 +370,7 @@ def write_objects(
@h5check_property_exists("objects")
def write_properties(
- self, data: Dict[str, Any], *, allow_overwrite: bool = False
+ self, data: dict[str, Any], *, allow_overwrite: bool = False
) -> None:
"""Write object properties to HDF file.
@@ -378,7 +390,7 @@ def write_properties(
grp = self._hdf[f"objects/{self.object_type}"]
- if "properties" not in grp.keys():
+ if "properties" not in grp:
props_grp = grp.create_group("properties")
else:
props_grp = self._hdf[f"objects/{self.object_type}/properties"]
@@ -395,31 +407,23 @@ def write_properties(
# Check if the property is already in the props_grp:
if key in props_grp:
- if allow_overwrite is False:
- logger.info(
- f"Property '{key}' already written in the file"
- )
+ if allow_overwrite:
+ del self._hdf[f"objects/{self.object_type}/properties"][key]
+ logger.info(f"Property '{key}' erased to be overwritten...")
+
+ else:
+ logger.info(f"Property '{key}' already written in the file")
raise KeyError(
f"Property '{key}' already in file -> switch on "
"'overwrite' param to replace existing property "
)
- else:
- del self._hdf[f"objects/{self.object_type}/properties"][
- key
- ]
- logger.info(
- f"Property '{key}' erased to be overwritten..."
- )
-
# Now that you handled overwriting, write the values:
- logger.info(
- f"Writing properties/{self.object_type}/{key} {values.shape}"
- )
+ logger.info(f"Writing properties/{self.object_type}/{key} {values.shape}")
props_grp.create_dataset(key, data=data[key], dtype="float32")
@property # type: ignore
@h5check_property_exists("tracks")
- def tracks(self) -> List[btypes.Tracklet]:
+ def tracks(self) -> list[btypes.Tracklet]:
"""Return the tracks in the file."""
logger.info(f"Loading tracks/{self.object_type}")
@@ -449,10 +453,9 @@ def tracks(self) -> List[btypes.Tracklet]:
obj = self.filtered_objects(f_expr=f_expr)
- def _get_txyz(_ref: int) -> int:
- if _ref >= 0:
- return obj[_ref]
- return dummy_obj[abs(_ref) - 1] # references are -ve for dummies
+ def _get_txyz(_ref: int) -> btypes.PyTrackObject:
+ # references are -ve for dummies
+ return obj[_ref] if _ref >= 0 else dummy_obj[abs(_ref) - 1]
tracks = []
for i in range(track_map.shape[0]):
@@ -467,7 +470,7 @@ def _get_txyz(_ref: int) -> int:
tracks.append(track)
# once we have all of the tracks, populate the children
- to_update = {}
+ to_update: dict = {}
for track in tracks:
if not track.is_root:
parents = filter(lambda t: track.parent == t.ID, tracks)
@@ -478,9 +481,7 @@ def _get_txyz(_ref: int) -> int:
# sanity check, can be removed at a later date
MAX_N_CHILDREN = 2
- assert all(
- len(children) <= MAX_N_CHILDREN for children in to_update.values()
- )
+ assert all(len(children) <= MAX_N_CHILDREN for children in to_update.values())
# add the children to the parent
for track, children in to_update.items():
@@ -490,7 +491,7 @@ def _get_txyz(_ref: int) -> int:
def write_tracks( # noqa: PLR0912
self,
- data: Union[List[btypes.Tracklet], BayesianTracker],
+ data: Union[list[btypes.Tracklet], BayesianTracker],
*,
f_expr: Optional[str] = None,
) -> None:
@@ -509,8 +510,8 @@ def write_tracks( # noqa: PLR0912
if not check_track_type(data):
raise ValueError(f"Data of type {type(data)} not supported.")
- all_objects = itertools.chain.from_iterable(
- [trk._data for trk in data]
+ all_objects = list(
+ itertools.chain.from_iterable([trk._data for trk in data])
)
objects = [obj for obj in all_objects if not obj.dummy]
@@ -590,7 +591,39 @@ def write_tracks( # noqa: PLR0912
@property # type: ignore
@h5check_property_exists("tracks")
- def lbep(self) -> np.ndarray:
+ def lbep(self) -> npt.NDArray:
"""Return the LBEP data."""
logger.info(f"Loading LBEP/{self.object_type}")
return self._hdf["tracks"][self.object_type]["LBEPR"][:]
+
+ def tree(self) -> None:
+ """Recursively iterate over the H5 file to reveal the tree structure and number
+ of elements within."""
+ _h5_tree(self._hdf)
+
+
+def _h5_tree(hdf, *, prefix: str = "") -> None:
+ """Recursively iterate over an H5 file to reveal the tree structure and number
+ of elements within. Writes the output to the default logger.
+
+ Parameters
+ ----------
+ hdf : hdf object
+ The hdf object to iterate over
+ prefix : str
+ A prepended string for layout
+ """
+ n_items = len(hdf)
+ for idx, (key, val) in enumerate(hdf.items()):
+ if idx == (n_items - 1):
+ # the last item
+ if isinstance(val, h5py._hl.group.Group):
+ logger.info(f"{prefix}└── {key}")
+ _h5_tree(val, prefix=f"{prefix} ")
+ else:
+ logger.info(f"{prefix}└── {key} ({len(val)})")
+ elif isinstance(val, h5py._hl.group.Group):
+ logger.info(f"{prefix}├── {key}")
+ _h5_tree(val, prefix=f"{prefix}│ ")
+ else:
+ logger.info(f"{prefix}├── {key} ({len(val)})")
diff --git a/btrack/io/importers.py b/btrack/io/importers.py
index a6b349f8..d0b34029 100644
--- a/btrack/io/importers.py
+++ b/btrack/io/importers.py
@@ -2,12 +2,11 @@
import csv
import os
-from typing import List
from btrack import btypes
-def import_CSV(filename: os.PathLike) -> List[btypes.PyTrackObject]:
+def import_CSV(filename: os.PathLike) -> list[btypes.PyTrackObject]:
"""Import localizations from a CSV file.
Parameters
@@ -17,7 +16,7 @@ def import_CSV(filename: os.PathLike) -> List[btypes.PyTrackObject]:
Returns
-------
- objects : List[btypes.PyTrackObject]
+ objects : list[btypes.PyTrackObject]
A list of objects in the CSV file.
Notes
@@ -45,7 +44,7 @@ def import_CSV(filename: os.PathLike) -> List[btypes.PyTrackObject]:
csvreader = csv.DictReader(csv_file, delimiter=",", quotechar="|")
for i, row in enumerate(csvreader):
data = {k: float(v) for k, v in row.items()}
- data.update({"ID": i})
+ data["ID"] = i
obj = btypes.PyTrackObject.from_dict(data)
objects.append(obj)
return objects
diff --git a/btrack/io/utils.py b/btrack/io/utils.py
index 60aaba2c..948f64df 100644
--- a/btrack/io/utils.py
+++ b/btrack/io/utils.py
@@ -1,9 +1,10 @@
from __future__ import annotations
import logging
-from typing import Any, Dict, List, Union
+from typing import Any, Union
import numpy as np
+from numpy import typing as npt
# import core
from btrack import btypes, constants
@@ -13,15 +14,13 @@
def localizations_to_objects(
- localizations: Union[
- np.ndarray, List[btypes.PyTrackObject], Dict[str, Any]
- ]
-) -> List[btypes.PyTrackObject]:
+ localizations: Union[npt.NDArray, list[btypes.PyTrackObject], dict[str, Any]]
+) -> list[btypes.PyTrackObject]:
"""Take a numpy array or pandas dataframe and convert to PyTrackObjects.
Parameters
----------
- localizations : list[PyTrackObject], np.ndarray, pandas.DataFrame
+ localizations : list[PyTrackObject], npt.NDArray, pandas.DataFrame
A list or array of localizations.
Returns
@@ -39,16 +38,11 @@ def localizations_to_objects(
# do we have a numpy array or pandas dataframe?
if isinstance(localizations, np.ndarray):
return objects_from_array(localizations)
- else:
- try:
- objects_dict = {
- c: np.asarray(localizations[c]) for c in localizations
- }
- except ValueError as err:
- logger.error(f"Unknown localization type: {type(localizations)}")
- raise TypeError(
- f"Unknown localization type: {type(localizations)}"
- ) from err
+ try:
+ objects_dict = {c: np.asarray(localizations[c]) for c in localizations}
+ except ValueError as err:
+ logger.error(f"Unknown localization type: {type(localizations)}")
+ raise TypeError(f"Unknown localization type: {type(localizations)}") from err
# how many objects are there
n_objects = objects_dict["t"].shape[0]
@@ -57,7 +51,7 @@ def localizations_to_objects(
return objects_from_dict(objects_dict)
-def objects_from_dict(objects_dict: dict) -> List[btypes.PyTrackObject]:
+def objects_from_dict(objects_dict: dict) -> list[btypes.PyTrackObject]:
"""Construct PyTrackObjects from a dictionary"""
# now that we have the object dictionary, convert this to objects
objects = []
@@ -73,10 +67,10 @@ def objects_from_dict(objects_dict: dict) -> List[btypes.PyTrackObject]:
def objects_from_array(
- objects_arr: np.ndarray,
+ objects_arr: npt.NDArray,
*,
- default_keys: List[str] = constants.DEFAULT_OBJECT_KEYS,
-) -> List[btypes.PyTrackObject]:
+ default_keys: list[str] = constants.DEFAULT_OBJECT_KEYS,
+) -> list[btypes.PyTrackObject]:
"""Construct PyTrackObjects from a numpy array."""
assert objects_arr.ndim == constants.Dimensionality.TWO
diff --git a/btrack/libwrapper.py b/btrack/libwrapper.py
index 8b3911c7..e1c84f07 100644
--- a/btrack/libwrapper.py
+++ b/btrack/libwrapper.py
@@ -23,25 +23,19 @@ def numpy_pointer_decorator(func):
@numpy_pointer_decorator
def np_dbl_p():
"""Temporary function. Will remove in final release"""
- return np.ctypeslib.ndpointer(
- dtype=np.double, ndim=2, flags="C_CONTIGUOUS"
- )
+ return np.ctypeslib.ndpointer(dtype=np.double, ndim=2, flags="C_CONTIGUOUS")
@numpy_pointer_decorator
def np_dbl_pc():
"""Temporary function. Will remove in final release"""
- return np.ctypeslib.ndpointer(
- dtype=np.double, ndim=2, flags="F_CONTIGUOUS"
- )
+ return np.ctypeslib.ndpointer(dtype=np.double, ndim=2, flags="F_CONTIGUOUS")
@numpy_pointer_decorator
def np_uint_p():
"""Temporary function. Will remove in final release"""
- return np.ctypeslib.ndpointer(
- dtype=np.uint32, ndim=2, flags="C_CONTIGUOUS"
- )
+ return np.ctypeslib.ndpointer(dtype=np.uint32, ndim=2, flags="C_CONTIGUOUS")
@numpy_pointer_decorator
@@ -53,9 +47,7 @@ def np_int_p():
@numpy_pointer_decorator
def np_int_vec_p():
"""Temporary function. Will remove in final release"""
- return np.ctypeslib.ndpointer(
- dtype=np.int32, ndim=1
- ) # , flags='C_CONTIGUOUS')
+ return np.ctypeslib.ndpointer(dtype=np.int32, ndim=1) # , flags='C_CONTIGUOUS')
@log_debug_info
diff --git a/btrack/models.py b/btrack/models.py
index f780f2af..ad70bf51 100644
--- a/btrack/models.py
+++ b/btrack/models.py
@@ -1,6 +1,7 @@
-from typing import List, Optional
+from typing import Optional
import numpy as np
+from numpy import typing as npt
from pydantic import BaseModel, root_validator, validator
from . import constants
@@ -9,9 +10,7 @@
__all__ = ["MotionModel", "ObjectModel", "HypothesisModel"]
-def _check_symmetric(
- x: np.ndarray, rtol: float = 1e-5, atol: float = 1e-8
-) -> bool:
+def _check_symmetric(x: npt.NDArray, rtol: float = 1e-5, atol: float = 1e-8) -> bool:
"""Check that a matrix is symmetric by comparing with it's own transpose."""
return np.allclose(x, x.T, rtol=rtol, atol=atol)
@@ -80,12 +79,12 @@ class MotionModel(BaseModel):
measurements: int
states: int
- A: np.ndarray
- H: np.ndarray
- P: np.ndarray
- R: np.ndarray
- G: Optional[np.ndarray] = None
- Q: Optional[np.ndarray] = None
+ A: npt.NDArray
+ H: npt.NDArray
+ P: npt.NDArray
+ R: npt.NDArray
+ G: Optional[npt.NDArray] = None
+ Q: Optional[npt.NDArray] = None
dt: float = 1.0
accuracy: float = 2.0
max_lost: int = constants.MAX_LOST
@@ -175,9 +174,9 @@ class ObjectModel(BaseModel):
"""
states: int
- emission: np.ndarray
- transition: np.ndarray
- start: np.ndarray
+ emission: npt.NDArray
+ transition: npt.NDArray
+ start: npt.NDArray
name: str = "Default"
@validator("emission", "transition", "start", pre=True)
@@ -259,7 +258,7 @@ class HypothesisModel(BaseModel):
.. math:: e^{(-d / \lambda)}
"""
- hypotheses: List[str]
+ hypotheses: list[str]
lambda_time: float
lambda_dist: float
lambda_link: float
@@ -277,15 +276,13 @@ class HypothesisModel(BaseModel):
@validator("hypotheses", pre=True)
def parse_hypotheses(cls, hypotheses):
- if not all(h in H_TYPES for h in hypotheses):
+ if any(h not in H_TYPES for h in hypotheses):
raise ValueError("Unknown hypothesis type in `hypotheses`.")
return hypotheses
def hypotheses_to_generate(self) -> int:
"""Return an integer representation of the hypotheses to generate."""
- h_bin = "".join(
- [str(int(h)) for h in [h in self.hypotheses for h in H_TYPES]]
- )
+ h_bin = "".join([str(int(h)) for h in [h in self.hypotheses for h in H_TYPES]])
return int(h_bin[::-1], 2)
def as_ctype(self) -> PyHypothesisParams:
diff --git a/btrack/napari.yaml b/btrack/napari.yaml
index 990d8be5..a74342e3 100644
--- a/btrack/napari.yaml
+++ b/btrack/napari.yaml
@@ -7,6 +7,9 @@ contributions:
- id: btrack.read_btrack
title: Read btrack files
python_name: btrack.napari.reader:get_reader
+ - id: btrack.write_hdf
+ title: Export Tracks to HDF
+ python_name: btrack.napari.writer:export_to_hdf
- id: btrack.track
title: Create Track
python_name: btrack.napari.main:create_btrack_widget
@@ -19,6 +22,11 @@ contributions:
- '*.hdf5'
accepts_directories: false
+ writers:
+ - command: btrack.write_hdf
+ layer_types: ["tracks"]
+ filename_extensions: [".h5", ".hdf", ".hdf5"]
+
widgets:
- command: btrack.track
display_name: Track
diff --git a/btrack/napari/__init__.py b/btrack/napari/__init__.py
index 92442fee..92ee399d 100644
--- a/btrack/napari/__init__.py
+++ b/btrack/napari/__init__.py
@@ -1,10 +1,3 @@
-try:
- from ._version import version as __version__
-except ImportError:
- __version__ = "unknown"
-
-import logging
-
from btrack.napari import constants, main
__all__ = [
diff --git a/btrack/napari/assets/btrack_logo.png b/btrack/napari/assets/btrack_logo.png
new file mode 100644
index 00000000..219dc507
Binary files /dev/null and b/btrack/napari/assets/btrack_logo.png differ
diff --git a/btrack/napari/config.py b/btrack/napari/config.py
index 7012edf8..fa5536e6 100644
--- a/btrack/napari/config.py
+++ b/btrack/napari/config.py
@@ -12,7 +12,7 @@
import numpy as np
import btrack
-from btrack import datasets
+import btrack.datasets
__all__ = [
"create_default_configs",
@@ -38,7 +38,7 @@ def __getitem__(self, matrix_name):
return self.__dict__[matrix_name]
def __setitem__(self, matrix_name, sigma):
- if matrix_name not in self.__dict__.keys():
+ if matrix_name not in self.__dict__:
_msg = f"Unknown matrix name '{matrix_name}'"
raise ValueError(_msg)
self.__dict__[matrix_name] = sigma
@@ -74,11 +74,10 @@ def __post_init__(self):
config = btrack.config.load_config(self.filename)
self.tracker_config, self.sigmas = self._unscale_config(config)
- def _unscale_config(
- self, config: TrackerConfig
- ) -> tuple[TrackerConfig, Sigmas]:
+ def _unscale_config(self, config: TrackerConfig) -> tuple[TrackerConfig, Sigmas]:
"""Convert the matrices of a scaled TrackerConfig MotionModel to unscaled."""
+ assert config.motion_model is not None
P_sigma = np.max(config.motion_model.P)
config.motion_model.P /= P_sigma
@@ -90,6 +89,7 @@ def _unscale_config(
# Instead, use G if it exists. If not, determine G from Q, which we can
# do because Q = G.T @ G
if config.motion_model.G is None:
+ assert config.motion_model.Q is not None
config.motion_model.G = config.motion_model.Q.diagonal() ** 0.5
G_sigma = np.max(config.motion_model.G)
config.motion_model.G /= G_sigma
@@ -107,8 +107,10 @@ def scale_config(self) -> TrackerConfig:
# Create a copy so that config values stay in sync with widget values
scaled_config = copy.deepcopy(self.tracker_config)
+ assert scaled_config.motion_model is not None
scaled_config.motion_model.P *= self.sigmas.P
scaled_config.motion_model.R *= self.sigmas.R
+ assert scaled_config.motion_model.G is not None
scaled_config.motion_model.G *= self.sigmas.G
scaled_config.motion_model.Q = (
scaled_config.motion_model.G.T @ scaled_config.motion_model.G
@@ -138,12 +140,12 @@ def __post_init__(self):
"""Add the default cell and particle configs."""
self.add_config(
- filename=datasets.cell_config(),
+ filename=btrack.datasets.cell_config(),
name="cell",
overwrite=False,
)
self.add_config(
- filename=datasets.particle_config(),
+ filename=btrack.datasets.particle_config(),
name="particle",
overwrite=False,
)
diff --git a/btrack/napari/constants.py b/btrack/napari/constants.py
index 668afed4..45a1b0db 100644
--- a/btrack/napari/constants.py
+++ b/btrack/napari/constants.py
@@ -3,16 +3,6 @@
napari plugin of the btrack package.
"""
-HYPOTHESES = [
- "P_FP",
- "P_init",
- "P_term",
- "P_link",
- "P_branch",
- "P_dead",
- "P_merge",
-]
-
HYPOTHESIS_SCALING_FACTORS = [
"lambda_time",
"lambda_dist",
@@ -26,5 +16,4 @@
"dist_thresh",
"time_thresh",
"apop_thresh",
- "relax",
]
diff --git a/btrack/napari/examples/show_btrack_widget.py b/btrack/napari/examples/show_btrack_widget.py
index 7c9c64de..5b32b9a7 100644
--- a/btrack/napari/examples/show_btrack_widget.py
+++ b/btrack/napari/examples/show_btrack_widget.py
@@ -15,15 +15,14 @@
napari.current_viewer()
_, btrack_widget = viewer.window.add_plugin_dock_widget(
- plugin_name="napari-btrack", widget_name="Track"
+ plugin_name="btrack", widget_name="Track"
)
segmentation = datasets.example_segmentation()
viewer.add_labels(segmentation)
# napari takes the first image layer as default anyway here, but better to be explicit
-btrack_widget.segmentation.choices = viewer.layers
-btrack_widget.segmentation.value = viewer.layers["segmentation"]
+btrack_widget.segmentation.setCurrentText(viewer.layers["segmentation"].name)
if __name__ == "__main__":
# The napari event loop needs to be run under here to allow the window
diff --git a/btrack/napari/main.py b/btrack/napari/main.py
index 3d34e412..6d6a5b3b 100644
--- a/btrack/napari/main.py
+++ b/btrack/napari/main.py
@@ -4,17 +4,14 @@
if TYPE_CHECKING:
import numpy.typing as npt
-
- from magicgui.widgets import Container
+ from qtpy import QtWidgets
from btrack.config import TrackerConfig
from btrack.napari.config import TrackerConfigs
import logging
+from pathlib import Path
-import qtpy.QtWidgets
-
-import magicgui.widgets
import napari
import btrack
@@ -44,90 +41,211 @@
logger.setLevel(logging.DEBUG)
-def create_btrack_widget() -> Container:
+def create_btrack_widget() -> btrack.napari.widgets.BtrackWidget:
"""Create widgets for the btrack plugin."""
# First create our UI along with some default configs for the widgets
all_configs = btrack.napari.config.create_default_configs()
- widgets = btrack.napari.widgets.create_widgets()
- btrack_widget = magicgui.widgets.Container(
- widgets=widgets, scrollable=True
+ btrack_widget = btrack.napari.widgets.BtrackWidget(
+ napari_viewer=napari.current_viewer(),
)
- btrack_widget.viewer = napari.current_viewer()
# Set the cell_config defaults in the gui
btrack.napari.sync.update_widgets_from_config(
unscaled_config=all_configs["cell"],
- container=btrack_widget,
+ btrack_widget=btrack_widget,
+ )
+
+ # Add any existing Labels layers to the segmentation selector
+ add_existing_labels(
+ viewer=btrack_widget.viewer,
+ combobox=btrack_widget.segmentation,
)
# Now set the callbacks
- btrack_widget.config.changed.connect(
+ btrack_widget.viewer.layers.events.inserted.connect(
+ lambda event: select_inserted_labels(
+ new_layer=event.value,
+ combobox=btrack_widget.segmentation,
+ ),
+ )
+
+ btrack_widget.viewer.layers.events.removed.connect(
+ lambda event: remove_deleted_labels(
+ deleted_layer=event.value,
+ combobox=btrack_widget.segmentation,
+ ),
+ )
+
+ btrack_widget.config_name.currentTextChanged.connect(
lambda selected: select_config(btrack_widget, all_configs, selected),
)
- btrack_widget.call_button.changed.connect(
+ # Disable the Optimiser tab if unchecked
+ for tab in range(btrack_widget._tabs.count()):
+ if btrack_widget._tabs.tabText(tab) == "Optimiser":
+ break
+ btrack_widget.enable_optimisation.toggled.connect(
+ lambda is_checked: btrack_widget._tabs.setTabEnabled(tab, is_checked)
+ )
+
+ btrack_widget.track_button.clicked.connect(
lambda: run(btrack_widget, all_configs),
)
- btrack_widget.reset_button.changed.connect(
+ btrack_widget.reset_button.clicked.connect(
lambda: restore_defaults(btrack_widget, all_configs),
)
- btrack_widget.save_config_button.changed.connect(
+ btrack_widget.save_config_button.clicked.connect(
lambda: save_config_to_json(btrack_widget, all_configs)
)
- btrack_widget.load_config_button.changed.connect(
+ btrack_widget.load_config_button.clicked.connect(
lambda: load_config_from_json(btrack_widget, all_configs)
)
- # there are lots of widgets so make the container scrollable
- scroll = qtpy.QtWidgets.QScrollArea()
- scroll.setWidget(btrack_widget._widget._qwidget)
- btrack_widget._widget._qwidget = scroll
-
return btrack_widget
+def add_existing_labels(
+ viewer: napari.Viewer,
+ combobox: QtWidgets.QComboBox,
+):
+ """Add all existing Labels layers in the viewer to a combobox"""
+
+ labels_layers = [
+ layer.name for layer in viewer.layers if isinstance(layer, napari.layers.Labels)
+ ]
+ combobox.addItems(labels_layers)
+
+
+def select_inserted_labels(
+ new_layer: napari.layers.Layer,
+ combobox: QtWidgets.QComboBox,
+):
+ """Update the selected Labels when a labels layer is added"""
+
+ if not isinstance(new_layer, napari.layers.Labels):
+ message = (
+ f"Not selecting new layer {new_layer.name} as input for the "
+ f"segmentation widget as {new_layer.name} is {type(new_layer)} "
+ "layer not an Labels layer."
+ )
+ logger.debug(message)
+ return
+
+ combobox.addItem(new_layer.name)
+ combobox.setCurrentText(new_layer.name)
+
+ # Update layer name when it changes
+ viewer = napari.current_viewer()
+ new_layer.events.name.connect(
+ lambda event: update_labels_name(
+ layer=event.source,
+ labels_layers=[
+ layer
+ for layer in viewer.layers
+ if isinstance(layer, napari.layers.Labels)
+ ],
+ combobox=combobox,
+ ),
+ )
+
+
+def update_labels_name(
+ layer: napari.layers.Layer,
+ labels_layers: list[napari.layer.Layer],
+ combobox: QtWidgets.QComboBox,
+):
+ """Update the name of an Labels layer"""
+
+ if not isinstance(layer, napari.layers.Labels):
+ message = (
+ f"Not updating name of layer {layer.name} as input for the "
+ f"segmentation widget as {layer.name} is {type(layer)} "
+ "layer not a Labels layer."
+ )
+ logger.debug(message)
+ return
+
+ layer_index = [layer.name for layer in labels_layers].index(layer.name)
+ combobox.setItemText(layer_index, layer.name)
+
+
+def remove_deleted_labels(
+ deleted_layer: napari.layers.Layer,
+ combobox: QtWidgets.QComboBox,
+):
+ """Remove the deleted Labels layer name from the combobox"""
+
+ if not isinstance(deleted_layer, napari.layers.Labels):
+ message = (
+ f"Not deleting layer {deleted_layer.name} from the segmentation "
+ f"widget as {deleted_layer.name} is {type(deleted_layer)} "
+ "layer not an Labels layer."
+ )
+ logger.debug(message)
+ return
+
+ layer_index = combobox.findText(deleted_layer.name)
+ combobox.removeItem(layer_index)
+
+
def select_config(
- btrack_widget: Container,
+ btrack_widget: btrack.napari.widgets.BtrackWidget,
configs: TrackerConfigs,
new_config_name: str,
) -> None:
"""Set widget values from a newly-selected base config"""
# first update the previous config with the current widget values
- previous_config_name = configs.current_config
- previous_config = configs[previous_config_name]
- previous_config = btrack.napari.sync.update_config_from_widgets(
- unscaled_config=previous_config,
- container=btrack_widget,
+ _ = btrack.napari.sync.update_config_from_widgets(
+ unscaled_config=configs[configs.current_config],
+ btrack_widget=btrack_widget,
)
# now load the newly-selected config and set widget values
configs.current_config = new_config_name
- new_config = configs[new_config_name]
- new_config = btrack.napari.sync.update_widgets_from_config(
- unscaled_config=new_config,
- container=btrack_widget,
+ _ = btrack.napari.sync.update_widgets_from_config(
+ unscaled_config=configs[new_config_name],
+ btrack_widget=btrack_widget,
)
-def run(btrack_widget: Container, configs: TrackerConfigs) -> None:
+def run(
+ btrack_widget: btrack.napari.widgets.BtrackWidget,
+ configs: TrackerConfigs,
+) -> None:
"""
Update the TrackerConfig from widget values, run tracking,
and add tracks to the viewer.
"""
- unscaled_config = configs[btrack_widget.config.current_choice]
+ # TODO:
+ # This method of showing the activity dock will be removed
+ # and replaced with a public method in the api
+ # See: https://github.com/napari/napari/issues/4598
+ activity_dock_visible = (
+ btrack_widget.viewer.window._qt_window._activity_dialog.isVisible()
+ )
+ btrack_widget.viewer.window._status_bar._toggle_activity_dock(visible=True)
+
+ if btrack_widget.segmentation.currentIndex() < 0:
+ napari.utils.notifications.show_error(
+ "No segmentation (Image layer) selected - cannot run tracking."
+ )
+ return
+
+ unscaled_config = configs[btrack_widget.config_name.currentText()]
unscaled_config = btrack.napari.sync.update_config_from_widgets(
unscaled_config=unscaled_config,
- container=btrack_widget,
+ btrack_widget=btrack_widget,
)
config = unscaled_config.scale_config()
- segmentation = btrack_widget.segmentation.value
+ segmentation_name = btrack_widget.segmentation.currentText()
+ segmentation = btrack_widget.viewer.layers[segmentation_name]
data, properties, graph = _run_tracker(segmentation, config)
btrack_widget.viewer.add_tracks(
@@ -135,8 +253,15 @@ def run(btrack_widget: Container, configs: TrackerConfigs) -> None:
properties=properties,
graph=graph,
name=f"{segmentation}_btrack",
+ scale=segmentation.scale,
+ translate=segmentation.translate,
)
+ btrack_widget.viewer.window._status_bar._toggle_activity_dock(activity_dock_visible)
+
+ message = f"Finished tracking for '{segmentation_name}'"
+ napari.utils.notifications.show_info(message)
+
def _run_tracker(
segmentation: napari.layers.Image | napari.layers.Labels,
@@ -145,11 +270,19 @@ def _run_tracker(
"""
Runs BayesianTracker with given segmentation and configuration.
"""
- with btrack.BayesianTracker() as tracker:
+ num_steps = 5 if tracker_config.enable_optimisation else 4
+
+ with btrack.BayesianTracker() as tracker, napari.utils.progress(
+ total=num_steps
+ ) as pbr:
+ pbr.set_description("Initialising the tracker")
tracker.configure(tracker_config)
+ pbr.update(1)
# append the objects to be tracked
+ pbr.set_description("Convert segmentation to trackable objects")
segmented_objects = segmentation_to_objects(segmentation.data)
+ pbr.update(1)
tracker.append(segmented_objects)
# set the volume
@@ -157,23 +290,30 @@ def _run_tracker(
# napari order of dimensions is T(Z)XY
# so we ignore the first dimension (time) and reverse the others
dimensions = segmentation.level_shapes[0, 1:]
- tracker.volume = tuple(
- (0, dimension) for dimension in reversed(dimensions)
- )
+ tracker.volume = tuple((0, dimension) for dimension in reversed(dimensions))
# track them (in interactive mode)
- tracker.track_interactive(step_size=100)
+ pbr.set_description("Run tracking")
+ tracker.track(step_size=100)
+ pbr.update(1)
- # generate hypotheses and run the global optimizer
- tracker.optimize()
+ if tracker.enable_optimisation:
+ # generate hypotheses and run the global optimizer
+ pbr.set_description("Run optimisation")
+ tracker.optimize()
+ pbr.update(1)
# get the tracks in a format for napari visualization
- data, properties, graph = tracker.to_napari(ndim=2)
+ pbr.set_description("Convert to napari tracks layer")
+ data, properties, graph = tracker.to_napari()
+ pbr.update(1)
+
return data, properties, graph
def restore_defaults(
- btrack_widget: Container, configs: TrackerConfigs
+ btrack_widget: btrack.napari.widgets.BtrackWidget,
+ configs: TrackerConfigs,
) -> None:
"""Reload the config file then set widgets to the config's default values."""
@@ -188,12 +328,13 @@ def restore_defaults(
config = configs[config_name]
config = btrack.napari.sync.update_widgets_from_config(
unscaled_config=config,
- container=btrack_widget,
+ btrack_widget=btrack_widget,
)
def save_config_to_json(
- btrack_widget: Container, configs: TrackerConfigs
+ btrack_widget: btrack.napari.widgets.BtrackWidget,
+ configs: TrackerConfigs,
) -> None:
"""Save widget values to file"""
@@ -206,18 +347,24 @@ def save_config_to_json(
logger.info(_msg)
return
- unscaled_config = configs[btrack_widget.config.current_choice]
+ unscaled_config = configs[btrack_widget.config_name.currentText()]
btrack.napari.sync.update_config_from_widgets(
unscaled_config=unscaled_config,
- container=btrack_widget,
+ btrack_widget=btrack_widget,
)
config = unscaled_config.scale_config()
+ # set the config name to match the filename
+ config.name = Path(save_path).stem
+ config.hypothesis_model.name = config.name
+ config.motion_model.name = config.name
+
btrack.config.save_config(save_path, config)
def load_config_from_json(
- btrack_widget: Container, configs: TrackerConfigs
+ btrack_widget: btrack.napari.widgets.BtrackWidget,
+ configs: TrackerConfigs,
) -> None:
"""Load a config from file and set it as the selected base config"""
@@ -228,6 +375,5 @@ def load_config_from_json(
return
config_name = configs.add_config(filename=load_path, overwrite=False)
- btrack_widget.config.options["choices"].append(config_name)
- btrack_widget.config.reset_choices()
- btrack_widget.config.value = config_name
+ btrack_widget.config_name.addItem(config_name)
+ btrack_widget.config_name.setCurrentText(config_name)
diff --git a/btrack/napari/reader.py b/btrack/napari/reader.py
index 65ced645..ad7559c1 100644
--- a/btrack/napari/reader.py
+++ b/btrack/napari/reader.py
@@ -2,9 +2,9 @@
This module is a reader plugin btrack files for napari.
"""
import os
-from typing import Callable, List, Optional, Sequence, Union
-
-from napari_plugin_engine import napari_hook_implementation
+import pathlib
+from collections.abc import Sequence
+from typing import Callable, Optional, Union
from napari.types import LayerDataTuple
@@ -13,10 +13,9 @@
# Type definitions
PathOrPaths = Union[os.PathLike, Sequence[os.PathLike]]
-ReaderFunction = Callable[[PathOrPaths], List[LayerDataTuple]]
+ReaderFunction = Callable[[PathOrPaths], list[LayerDataTuple]]
-@napari_hook_implementation
def get_reader(path: PathOrPaths) -> Optional[ReaderFunction]:
"""A basic implementation of the napari_get_reader hook specification.
@@ -31,10 +30,24 @@ def get_reader(path: PathOrPaths) -> Optional[ReaderFunction]:
If the path is a recognized format, return a function that accepts the
same path or list of paths, and returns a list of layer data tuples.
"""
- return reader_function
-
-
-def reader_function(path: PathOrPaths) -> List[LayerDataTuple]:
+ if isinstance(path, list):
+ # reader plugins may be handed single path, or a list of paths.
+ # if it is a list, it is assumed to be an image stack...
+ # so we are only going to look at the first file.
+ path = path[0]
+
+ # if we know we cannot read the file, we immediately return None.
+ supported_extensions = [
+ ".h5",
+ ".hdf",
+ ".hdf5",
+ ]
+ return (
+ reader_function if pathlib.Path(path).suffix in supported_extensions else None
+ )
+
+
+def reader_function(path: PathOrPaths) -> list[LayerDataTuple]:
"""Take a path or list of paths and return a list of LayerData tuples.
Readers are expected to return data as a list of tuples, where each tuple
@@ -57,10 +70,10 @@ def reader_function(path: PathOrPaths) -> List[LayerDataTuple]:
to layer_type=="image" if not provided
"""
# handle both a string and a list of strings
- paths = [path] if not isinstance(path, list) else path
+ paths = path if isinstance(path, list) else [path]
# store the layers to be generated
- layers: List[tuple] = []
+ layers: list[tuple] = []
for _path in paths:
with HDF5FileHandler(_path, "r") as hdf:
diff --git a/btrack/napari/sync.py b/btrack/napari/sync.py
index 48621ddd..73eb0011 100644
--- a/btrack/napari/sync.py
+++ b/btrack/napari/sync.py
@@ -7,59 +7,66 @@
from typing import TYPE_CHECKING
if TYPE_CHECKING:
- from magicgui.widgets import Container
-
- from btrack.config import TrackerConfig
+ import btrack.napari.widgets
from btrack.napari.config import Sigmas, UnscaledTrackerConfig
+from qtpy import QtCore
+
import btrack.napari.constants
def update_config_from_widgets(
unscaled_config: UnscaledTrackerConfig,
- container: Container,
-) -> TrackerConfig:
+ btrack_widget: btrack.napari.widgets.BtrackWidget,
+) -> UnscaledTrackerConfig:
"""Update an UnscaledTrackerConfig with the current widget values."""
-
- # Update MotionModel matrix scaling factors
- sigmas: Sigmas = unscaled_config.sigmas
- for matrix_name in sigmas:
- sigmas[matrix_name] = container[f"{matrix_name}_sigma"].value
-
- # Update TrackerConfig values
+ ## Retrieve model configs
config = unscaled_config.tracker_config
- update_method_name = container.update_method.current_choice
- update_method_index = container.update_method.choices.index(
- update_method_name
+ motion_model = config.motion_model
+ hypothesis_model = config.hypothesis_model
+
+ ## Update widgets from the Method tab
+ config.update_method = btrack_widget.update_method.currentIndex()
+ config.max_search_radius = btrack_widget.max_search_radius.value()
+ motion_model.max_lost = btrack_widget.max_lost.value()
+ motion_model.prob_not_assign = btrack_widget.prob_not_assign.value()
+ config.enable_optimisation = (
+ btrack_widget.enable_optimisation.checkState() == QtCore.Qt.CheckState.Checked
)
- config.update_method = update_method_index
- config.max_search_radius = container.max_search_radius.value
- # Update MotionModel values
- motion_model = config.motion_model
- motion_model.accuracy = container.accuracy.value
- motion_model.max_lost = container.max_lost.value
+ ## Update widgets from the Motion tab
+ sigmas: Sigmas = unscaled_config.sigmas
+ for matrix_name in sigmas:
+ sigmas[matrix_name] = btrack_widget[f"{matrix_name}_sigma"].value()
+ motion_model.accuracy = btrack_widget.accuracy.value()
- # Update HypothesisModel.hypotheses values
- hypothesis_model = config.hypothesis_model
+ ## Update widgets from the Optimiser tab
+ # HypothesisModel.hypotheses values
hypothesis_model.hypotheses = [
hypothesis
- for hypothesis in btrack.napari.constants.HYPOTHESES
- if container[hypothesis].value
+ for i, hypothesis in enumerate(btrack.optimise.hypothesis.H_TYPES)
+ if btrack_widget["hypotheses"].item(i).checkState()
+ == QtCore.Qt.CheckState.Checked
]
- # Update HypothesisModel scaling factors
+ # HypothesisModel scaling factors
for scaling_factor in btrack.napari.constants.HYPOTHESIS_SCALING_FACTORS:
setattr(
- hypothesis_model, scaling_factor, container[scaling_factor].value
+ hypothesis_model,
+ scaling_factor,
+ btrack_widget[scaling_factor].value(),
)
- # Update HypothesisModel thresholds
+ # HypothesisModel thresholds
for threshold in btrack.napari.constants.HYPOTHESIS_THRESHOLDS:
- setattr(hypothesis_model, threshold, container[threshold].value)
+ setattr(hypothesis_model, threshold, btrack_widget[threshold].value())
+ # other
hypothesis_model.segmentation_miss_rate = (
- container.segmentation_miss_rate.value
+ btrack_widget.segmentation_miss_rate.value()
+ )
+ hypothesis_model.relax = (
+ btrack_widget.relax.checkState() == QtCore.Qt.CheckState.Checked
)
return unscaled_config
@@ -67,46 +74,54 @@ def update_config_from_widgets(
def update_widgets_from_config(
unscaled_config: UnscaledTrackerConfig,
- container: Container,
-) -> Container:
+ btrack_widget: btrack.napari.widgets.BtrackWidget,
+) -> btrack.napari.widgets.BtrackWidget:
"""
- Update the widgets in a container with the values in an
+ Update the widgets in a btrack_widget with the values in an
UnscaledTrackerConfig.
"""
-
- # Update widgets from MotionModel matrix scaling factors
- sigmas: Sigmas = unscaled_config.sigmas
- for matrix_name in sigmas:
- container[f"{matrix_name}_sigma"].value = sigmas[matrix_name]
-
- # Update widgets from TrackerConfig values
+ ## Retrieve model configs
config = unscaled_config.tracker_config
- container.update_method.value = config.update_method.name
- container.max_search_radius.value = config.max_search_radius
-
- # Update widgets from MotionModel values
motion_model = config.motion_model
- container.accuracy.value = motion_model.accuracy
- container.max_lost.value = motion_model.max_lost
-
- # Update widgets from HypothesisModel.hypotheses values
hypothesis_model = config.hypothesis_model
- for hypothesis in btrack.napari.constants.HYPOTHESES:
- is_checked = hypothesis in hypothesis_model.hypotheses
- container[hypothesis].value = is_checked
- # Update widgets from HypothesisModel scaling factors
- for scaling_factor in btrack.napari.constants.HYPOTHESIS_SCALING_FACTORS:
- container[scaling_factor].value = getattr(
- hypothesis_model, scaling_factor
+ ## Update widgets from the Method tab
+ btrack_widget.update_method.setCurrentText(config.update_method.name)
+ btrack_widget.max_search_radius.setValue(config.max_search_radius)
+ btrack_widget.max_lost.setValue(motion_model.max_lost)
+ btrack_widget.prob_not_assign.setValue(motion_model.prob_not_assign)
+ btrack_widget.enable_optimisation.setChecked(config.enable_optimisation)
+
+ ## Update widgets from the Motion tab
+ sigmas: Sigmas = unscaled_config.sigmas
+ for matrix_name in sigmas:
+ btrack_widget[f"{matrix_name}_sigma"].setValue(sigmas[matrix_name])
+ btrack_widget.accuracy.setValue(motion_model.accuracy)
+
+ ## Update widgets from the Optimiser tab
+ # HypothesisModel.hypotheses values
+ for i, hypothesis in enumerate(btrack.optimise.hypothesis.H_TYPES):
+ is_checked = (
+ QtCore.Qt.CheckState.Checked
+ if hypothesis in hypothesis_model.hypotheses
+ else QtCore.Qt.CheckState.Unchecked
)
+ btrack_widget["hypotheses"].item(i).setCheckState(is_checked)
+
+ # HypothesisModel scaling factors
+ for scaling_factor in btrack.napari.constants.HYPOTHESIS_SCALING_FACTORS:
+ new_value = getattr(hypothesis_model, scaling_factor)
+ btrack_widget[scaling_factor].setValue(new_value)
- # Update widgets from HypothesisModel thresholds
+ # HypothesisModel thresholds
for threshold in btrack.napari.constants.HYPOTHESIS_THRESHOLDS:
- container[threshold].value = getattr(hypothesis_model, threshold)
+ new_value = getattr(hypothesis_model, threshold)
+ btrack_widget[threshold].setValue(new_value)
- container.segmentation_miss_rate.value = (
+ # other
+ btrack_widget.segmentation_miss_rate.setValue(
hypothesis_model.segmentation_miss_rate
)
+ btrack_widget.relax.setChecked(hypothesis_model.relax)
- return container
+ return btrack_widget
diff --git a/btrack/napari/widgets/__init__.py b/btrack/napari/widgets/__init__.py
index b14a892c..192a0d95 100644
--- a/btrack/napari/widgets/__init__.py
+++ b/btrack/napari/widgets/__init__.py
@@ -1,4 +1,4 @@
-from btrack.napari.widgets.create_ui import create_widgets
+from btrack.napari.widgets.create_ui import BtrackWidget
from btrack.napari.widgets.io import (
load_path_dialogue_box,
save_path_dialogue_box,
diff --git a/btrack/napari/widgets/_general.py b/btrack/napari/widgets/_general.py
index 3bd17be8..785db97f 100644
--- a/btrack/napari/widgets/_general.py
+++ b/btrack/napari/widgets/_general.py
@@ -1,84 +1,123 @@
from __future__ import annotations
-from typing import TYPE_CHECKING
+from pathlib import Path
-if TYPE_CHECKING:
- from magicgui.widgets import Widget
+from qtpy import QtCore, QtGui, QtWidgets
-import magicgui
-import napari
+def create_logo_widgets() -> dict[str, QtWidgets.QWidget]:
+ """Creates the widgets for the title, logo and documentation"""
-def create_input_widgets() -> list[Widget]:
+ title = QtWidgets.QLabel("Bayesian Tracker
")
+ title.setAlignment(QtCore.Qt.AlignHCenter)
+ widgets = {"title": title}
+
+ logo = QtWidgets.QLabel()
+ pixmap = QtGui.QPixmap(
+ str(Path(__file__).resolve().parents[1] / "assets" / "btrack_logo.png")
+ )
+ logo.setAlignment(QtCore.Qt.AlignHCenter)
+ scale = 0.8
+ logo.setPixmap(
+ pixmap.scaled(
+ int(pixmap.width() * scale),
+ int(pixmap.height() * scale),
+ QtCore.Qt.KeepAspectRatio,
+ )
+ )
+ widgets["logo"] = logo
+
+ docs = QtWidgets.QLabel('Documentation')
+ docs.setAlignment(QtCore.Qt.AlignHCenter)
+ docs.setOpenExternalLinks(True) # noqa: FBT003
+ docs.setTextFormat(QtCore.Qt.RichText)
+ docs.setTextInteractionFlags(QtCore.Qt.TextBrowserInteraction)
+ widgets["documentation"] = docs
+
+ return widgets
+
+
+def create_input_widgets() -> dict[str, tuple[str, QtWidgets.QWidget]]:
"""Create widgets for selecting labels layer and TrackerConfig"""
- segmentation_tooltip = (
+ segmentation = QtWidgets.QComboBox()
+ segmentation.setToolTip(
"Select a 'Labels' layer to use for tracking.\n"
"To use an 'Image' layer, first convert 'Labels' by right-clicking "
"on it in the layers list, and clicking on 'Convert to Labels'"
)
- segmentation = magicgui.widgets.create_widget(
- annotation=napari.layers.Labels,
- name="segmentation",
- label="segmentation",
- options={"tooltip": segmentation_tooltip},
- )
+ widgets = {"segmentation": ("segmentation", segmentation)}
- config_tooltip = (
- "Select a loaded configuration.\n"
- "Note, this will update values set below."
- )
- config = magicgui.widgets.create_widget(
- value="cell",
- name="config",
- label="config name",
- widget_type="ComboBox",
- options={
- "choices": ["cell", "particle"],
- "tooltip": config_tooltip,
- },
+ config_name = QtWidgets.QComboBox()
+ config_name.addItems(["cell", "particle"])
+ config_name.setToolTip(
+ "Select a loaded configuration.\nNote, this will update values set below."
)
+ widgets["config_name"] = ("config name", config_name)
- return [segmentation, config]
+ return widgets
-def create_update_method_widgets() -> list[Widget]:
+def create_basic_widgets() -> dict[str, tuple[str, QtWidgets.QWidget]]:
"""Create widgets for selecting the update method"""
- update_method_tooltip = (
+ update_method = QtWidgets.QComboBox()
+ update_method.addItems(
+ [
+ "EXACT",
+ "APPROXIMATE",
+ ]
+ )
+ update_method.setToolTip(
"Select the update method.\n"
"EXACT: exact calculation of Bayesian belief matrix.\n"
"APPROXIMATE: approximate the Bayesian belief matrix. Useful for datasets with "
"more than 1000 particles per frame."
)
- update_method = magicgui.widgets.create_widget(
- value="EXACT",
- name="update_method",
- label="update method",
- widget_type="ComboBox",
- options={
- "choices": ["EXACT", "APPROXIMATE"],
- "tooltip": update_method_tooltip,
- },
- )
+ widgets = {"update_method": ("update method", update_method)}
- # TODO: this widget should be hidden when the update method is set to EXACT
- max_search_radius_tooltip = (
+ max_search_radius = QtWidgets.QDoubleSpinBox()
+ max_search_radius.setRange(0, 1000)
+ max_search_radius.setStepType(QtWidgets.QAbstractSpinBox.AdaptiveDecimalStepType)
+ max_search_radius.setToolTip(
"The local spatial search radius (isotropic, pixels) used when the update "
"method is 'APPROXIMATE'"
)
- max_search_radius = magicgui.widgets.create_widget(
- value=100,
- name="max_search_radius",
- label="search radius",
- widget_type="SpinBox",
- options={"tooltip": max_search_radius_tooltip},
+ max_search_radius.setWrapping(True) # noqa: FBT003
+ widgets["max_search_radius"] = ("search radius", max_search_radius)
+
+ max_lost_frames = QtWidgets.QSpinBox()
+ max_lost_frames.setRange(0, 10)
+ max_lost_frames.setStepType(QtWidgets.QAbstractSpinBox.AdaptiveDecimalStepType)
+ max_lost_frames.setToolTip(
+ "Number of frames without observation before marking as lost"
+ )
+ widgets["max_lost"] = ("max lost", max_lost_frames)
+
+ not_assign = QtWidgets.QDoubleSpinBox()
+ not_assign.setDecimals(3)
+ not_assign.setRange(0, 1)
+ not_assign.setStepType(QtWidgets.QAbstractSpinBox.AdaptiveDecimalStepType)
+ not_assign.setToolTip("Default probability to not assign a track")
+ widgets["prob_not_assign"] = (
+ "P(not track)",
+ not_assign,
+ )
+
+ optimise = QtWidgets.QCheckBox()
+ optimise.setChecked(True) # noqa: FBT003
+ optimise.setToolTip(
+ "Enable the track optimisation.\n"
+ "This means that tracks will be optimised using the hypotheses"
+ "specified in the optimiser tab."
)
+ optimise.setTristate(False) # noqa: FBT003
+ widgets["enable_optimisation"] = ("enable optimisation", optimise)
- return [update_method, max_search_radius]
+ return widgets
-def create_control_widgets() -> list[Widget]:
+def create_config_widgets() -> dict[str, QtWidgets.QWidget]:
"""Create widgets for running the analysis or handling I/O.
This includes widgets for running the tracking, saving and loading
@@ -89,29 +128,32 @@ def create_control_widgets() -> list[Widget]:
"load_config_button",
"save_config_button",
"reset_button",
- "call_button",
]
labels = [
- "Load configuration",
- "Save configuration",
- "Reset defaults",
- "Run",
+ "Load Configuration",
+ "Save Configuration",
+ "Reset Defaults",
]
tooltips = [
"Load a TrackerConfig json file.",
"Export the current configuration to a TrackerConfig json file.",
"Reset the current configuration to the defaults stored in the corresponding json file.", # noqa: E501
- "Run the tracking analysis with the current configuration.",
]
- control_buttons = []
+ widgets = {}
for name, label, tooltip in zip(names, labels, tooltips):
- widget = magicgui.widgets.create_widget(
- name=name,
- label=label,
- widget_type="PushButton",
- options={"tooltip": tooltip},
- )
- control_buttons.append(widget)
+ widget = QtWidgets.QPushButton()
+ widget.setText(label)
+ widget.setToolTip(tooltip)
+ widgets[name] = widget
+
+ return widgets
+
+
+def create_track_widgets() -> dict[str, QtWidgets.QWidget]:
+ track_button = QtWidgets.QPushButton("Track")
+ track_button.setToolTip(
+ "Run the tracking analysis with the current configuration.",
+ )
- return control_buttons
+ return {"track_button": track_button}
diff --git a/btrack/napari/widgets/_hypothesis.py b/btrack/napari/widgets/_hypothesis.py
deleted file mode 100644
index a8b441f8..00000000
--- a/btrack/napari/widgets/_hypothesis.py
+++ /dev/null
@@ -1,214 +0,0 @@
-from __future__ import annotations
-
-from typing import TYPE_CHECKING
-
-if TYPE_CHECKING:
- from magicgui.widgets import Widget
-
-import magicgui
-
-import btrack.napari.constants
-
-
-def _create_hypotheses_widgets() -> list[Widget]:
- """Create widgets for selecting which hypotheses to generate."""
-
- hypotheses = btrack.napari.constants.HYPOTHESES
- tooltips = [
- "Hypothesis that a tracklet is a false positive detection. Always required.",
- "Hypothesis that a tracklet starts at the beginning of the movie or edge of the field of view.", # noqa: E501
- "Hypothesis that a tracklet ends at the end of the movie or edge of the field of view.", # noqa: E501
- "Hypothesis that two tracklets should be linked together.",
- "Hypothesis that a tracklet can split into two daughter tracklets.",
- "Hypothesis that a tracklet terminates without leaving the field of view.",
- "Hypothesis that two tracklets merge into one tracklet.",
- ]
-
- hypotheses_widgets = []
- for hypothesis, tooltip in zip(hypotheses, tooltips):
- widget = magicgui.widgets.create_widget(
- value=True,
- name=hypothesis,
- label=hypothesis,
- widget_type="CheckBox",
- options={"tooltip": tooltip},
- )
- hypotheses_widgets.append(widget)
-
- # P_FP is always required
- P_FP_hypothesis = hypotheses_widgets[0]
- P_FP_hypothesis.enabled = False
-
- # P_merge should be disabled by default
- P_merge_hypothesis = hypotheses_widgets[-1]
- P_merge_hypothesis.value = False
-
- return hypotheses_widgets
-
-
-def _create_scaling_factor_widgets() -> list[Widget]:
- """Create widgets for setting the scaling factors of the HypothesisModel"""
-
- widget_values = [5.0, 3.0, 10.0, 50.0]
- names = [
- "lambda_time",
- "lambda_dist",
- "lambda_link",
- "lambda_branch",
- ]
- labels = [
- "λ time",
- "λ distance",
- "λ linking",
- "λ branching",
- ]
- tooltips = [
- "Scaling factor for the influence of time when determining initialization or termination hypotheses.", # noqa: E501
- "Scaling factor for the influence of distance at the border when determining initialization or termination hypotheses.", # noqa: E501
- "Scaling factor for the influence of track-to-track distance on linking probability.", # noqa: E501
- "Scaling factor for the influence of cell state and position on division (mitosis/branching) probability.", # noqa: E501
- ]
-
- scaling_factor_widgets = []
- for value, name, label, tooltip in zip(
- widget_values, names, labels, tooltips
- ):
- widget = magicgui.widgets.create_widget(
- value=value,
- name=name,
- label=label,
- widget_type="FloatSpinBox",
- options={"tooltip": tooltip},
- )
- scaling_factor_widgets.append(widget)
-
- return scaling_factor_widgets
-
-
-def _create_threshold_widgets() -> list[Widget]:
- """Create widgets for setting thresholds for the HypothesisModel"""
-
- distance_threshold_tooltip = (
- "A threshold distance from the edge of the field of view to add an "
- "initialization or termination hypothesis."
- )
- distance_threshold = magicgui.widgets.create_widget(
- value=20.0,
- name="theta_dist",
- label="distance threshold",
- widget_type="FloatSpinBox",
- options={"tooltip": distance_threshold_tooltip},
- )
-
- time_threshold_tooltip = (
- "A threshold time from the beginning or end of movie to add "
- "an initialization or termination hypothesis."
- )
- time_threshold = magicgui.widgets.create_widget(
- value=5.0,
- name="theta_time",
- label="time threshold",
- widget_type="FloatSpinBox",
- options={"tooltip": time_threshold_tooltip},
- )
-
- apoptosis_threshold_tooltip = (
- "Number of apoptotic detections to be considered a genuine event.\n"
- "Detections are counted consecutively from the back of the track"
- )
- apoptosis_threshold = magicgui.widgets.create_widget(
- value=5,
- name="apop_thresh",
- label="apoptosis threshold",
- widget_type="SpinBox",
- options={"tooltip": apoptosis_threshold_tooltip},
- )
-
- return [
- distance_threshold,
- time_threshold,
- apoptosis_threshold,
- ]
-
-
-def _create_bin_size_widgets() -> list[Widget]:
- """Create widget for setting bin sizes for the HypothesisModel"""
-
- distance_bin_size_tooltip = (
- "Isotropic spatial bin size for considering hypotheses.\n"
- "Larger bin sizes generate more hypothesese for each tracklet."
- )
- distance_bin_size = magicgui.widgets.create_widget(
- value=40.0,
- name="dist_thresh",
- label="distance bin size",
- widget_type="FloatSpinBox",
- options={"tooltip": distance_bin_size_tooltip},
- )
-
- time_bin_size_tooltip = (
- "Temporal bin size for considering hypotheses.\n"
- "Larger bin sizes generate more hypothesese for each tracklet."
- )
- time_bin_size = magicgui.widgets.create_widget(
- value=2.0,
- name="time_thresh",
- label="time bin size",
- widget_type="FloatSpinBox",
- options={"tooltip": time_bin_size_tooltip},
- )
-
- return [
- distance_bin_size,
- time_bin_size,
- ]
-
-
-def create_hypothesis_model_widgets() -> list[Widget]:
- """Create widgets for setting parameters of the MotionModel"""
-
- hypothesis_model_label = magicgui.widgets.create_widget(
- label="Hypothesis model", # bold label
- widget_type="Label",
- gui_only=True,
- )
-
- hypotheses_widgets = _create_hypotheses_widgets()
- scaling_factor_widgets = _create_scaling_factor_widgets()
- threshold_widgets = _create_threshold_widgets()
- bin_size_widgets = _create_bin_size_widgets()
-
- segmentation_miss_rate_tooltip = (
- "Miss rate for the segmentation.\n"
- "e.g. 1/100 segmentations incorrect gives a segmentation miss rate of 0.01."
- )
- segmentation_miss_rate = magicgui.widgets.create_widget(
- value=0.1,
- name="segmentation_miss_rate",
- label="miss rate",
- widget_type="FloatSpinBox",
- options={"tooltip": segmentation_miss_rate_tooltip},
- )
-
- relax_tooltip = (
- "Disable the time and distance thresholds.\n"
- "This means that tracks can initialize or terminate anywhere and"
- "at any time in the dataset."
- )
- relax = magicgui.widgets.create_widget(
- value=True,
- name="relax",
- label="relax thresholds",
- widget_type="CheckBox",
- options={"tooltip": relax_tooltip},
- )
-
- return [
- hypothesis_model_label,
- *hypotheses_widgets,
- *scaling_factor_widgets,
- *threshold_widgets,
- *bin_size_widgets,
- segmentation_miss_rate,
- relax,
- ]
diff --git a/btrack/napari/widgets/_motion.py b/btrack/napari/widgets/_motion.py
index 3000fb5d..a4f1893b 100644
--- a/btrack/napari/widgets/_motion.py
+++ b/btrack/napari/widgets/_motion.py
@@ -1,97 +1,45 @@
from __future__ import annotations
-from typing import TYPE_CHECKING
+from qtpy import QtWidgets
-if TYPE_CHECKING:
- from magicgui.widgets import Widget
-import magicgui
-
-
-def _make_label_bold(label: str) -> str:
- """Generate html for a bold label"""
-
- return f"{label}"
-
-
-def _create_sigma_widgets() -> list[Widget]:
+def _create_sigma_widgets() -> dict[str, tuple[str, QtWidgets.QWidget]]:
"""Create widgets for setting the magnitudes of the MotionModel matrices"""
- P_sigma_tooltip = (
- "Magnitude of error in initial estimates.\n"
- "Used to scale the matrix P."
- )
- P_sigma = magicgui.widgets.create_widget(
- value=150.0,
- name="P_sigma",
- label=f"max({_make_label_bold('P')})",
- widget_type="FloatSpinBox",
- options={"tooltip": P_sigma_tooltip},
+ P_sigma = QtWidgets.QDoubleSpinBox()
+ P_sigma.setRange(0, 500)
+ P_sigma.setStepType(QtWidgets.QAbstractSpinBox.AdaptiveDecimalStepType)
+ P_sigma.setToolTip(
+ "Magnitude of error in initial estimates.\nUsed to scale the matrix P."
)
+ widgets = {"P_sigma": ("max(P)", P_sigma)}
- G_sigma_tooltip = (
- "Magnitude of error in process.\n Used to scale the matrix G."
- )
- G_sigma = magicgui.widgets.create_widget(
- value=15.0,
- name="G_sigma",
- label=f"max({_make_label_bold('G')})",
- widget_type="FloatSpinBox",
- options={"tooltip": G_sigma_tooltip},
- )
+ G_sigma = QtWidgets.QDoubleSpinBox()
+ G_sigma.setRange(0, 500)
+ G_sigma.setStepType(QtWidgets.QAbstractSpinBox.AdaptiveDecimalStepType)
+ G_sigma.setToolTip("Magnitude of error in process\nUsed to scale the matrix G.")
+ widgets["G_sigma"] = ("max(G)", G_sigma)
- R_sigma_tooltip = (
- "Magnitude of error in measurements.\n Used to scale the matrix R."
- )
- R_sigma = magicgui.widgets.create_widget(
- value=5.0,
- name="R_sigma",
- label=f"max({_make_label_bold('R')})",
- widget_type="FloatSpinBox",
- options={"tooltip": R_sigma_tooltip},
+ R_sigma = QtWidgets.QDoubleSpinBox()
+ R_sigma.setRange(0, 500)
+ R_sigma.setStepType(QtWidgets.QAbstractSpinBox.AdaptiveDecimalStepType)
+ R_sigma.setToolTip(
+ "Magnitude of error in measurements.\nUsed to scale the matrix R."
)
+ widgets["R_sigma"] = ("max(R)", R_sigma)
- return [
- P_sigma,
- G_sigma,
- R_sigma,
- ]
+ return widgets
-def create_motion_model_widgets() -> list[Widget]:
+def create_motion_model_widgets() -> dict[str, tuple[str, QtWidgets.QWidget]]:
"""Create widgets for setting parameters of the MotionModel"""
- motion_model_label = magicgui.widgets.create_widget(
- label=_make_label_bold("Motion model"),
- widget_type="Label",
- gui_only=True,
- )
-
- sigma_widgets = _create_sigma_widgets()
+ widgets = _create_sigma_widgets()
- accuracy_tooltip = "Integration limits for calculating probabilities"
- accuracy = magicgui.widgets.create_widget(
- value=7.5,
- name="accuracy",
- label="accuracy",
- widget_type="FloatSpinBox",
- options={"tooltip": accuracy_tooltip},
- )
-
- max_lost_frames_tooltip = (
- "Number of frames without observation before marking as lost"
- )
- max_lost_frames = magicgui.widgets.create_widget(
- value=5,
- name="max_lost",
- label="max lost",
- widget_type="SpinBox",
- options={"tooltip": max_lost_frames_tooltip},
- )
+ accuracy = QtWidgets.QDoubleSpinBox()
+ accuracy.setRange(0.1, 10)
+ accuracy.setStepType(QtWidgets.QAbstractSpinBox.AdaptiveDecimalStepType)
+ accuracy.setToolTip("Integration limits for calculating probabilities")
+ widgets["accuracy"] = ("accuracy", accuracy)
- return [
- motion_model_label,
- *sigma_widgets,
- accuracy,
- max_lost_frames,
- ]
+ return widgets
diff --git a/btrack/napari/widgets/_optimiser.py b/btrack/napari/widgets/_optimiser.py
new file mode 100644
index 00000000..f3658b3f
--- /dev/null
+++ b/btrack/napari/widgets/_optimiser.py
@@ -0,0 +1,145 @@
+from __future__ import annotations
+
+from qtpy import QtCore, QtWidgets
+
+import btrack.napari.constants
+
+
+def _create_hypotheses_widgets() -> dict[str, tuple[str, QtWidgets.QWidget]]:
+ """Create widgets for selecting which hypotheses to generate."""
+
+ hypotheses = btrack.optimise.hypothesis.H_TYPES
+ tooltips = [
+ "Hypothesis that a tracklet is a false positive detection. Always required.",
+ "Hypothesis that a tracklet starts at the beginning of the movie or edge of the field of view.", # noqa: E501
+ "Hypothesis that a tracklet ends at the end of the movie or edge of the field of view.", # noqa: E501
+ "Hypothesis that two tracklets should be linked together.",
+ "Hypothesis that a tracklet can split into two daughter tracklets.",
+ "Hypothesis that a tracklet terminates without leaving the field of view.",
+ "Hypothesis that two tracklets merge into one tracklet.",
+ ]
+
+ widget = QtWidgets.QListWidget()
+ widget.addItems([f"{h.replace('_', '(')})" for h in hypotheses])
+ flags = QtCore.Qt.ItemFlags(QtCore.Qt.ItemIsUserCheckable | QtCore.Qt.ItemIsEnabled)
+ for i, tooltip in enumerate(tooltips):
+ widget.item(i).setFlags(flags)
+ widget.item(i).setToolTip(tooltip)
+
+ # P_FP is always required
+ widget.item(hypotheses.index("P_FP")).setFlags(
+ QtCore.Qt.ItemIsUserCheckable,
+ )
+
+ return {"hypotheses": ("hypotheses", widget)}
+
+
+def _create_scaling_factor_widgets() -> dict[str, tuple[str, QtWidgets.QWidget]]:
+ """Create widgets for setting the scaling factors of the HypothesisModel"""
+
+ names = btrack.napari.constants.HYPOTHESIS_SCALING_FACTORS
+ labels = [
+ "λ time",
+ "λ distance",
+ "λ linking",
+ "λ branching",
+ ]
+ tooltips = [
+ "Scaling factor for the influence of time when determining initialization or termination hypotheses.", # noqa: E501
+ "Scaling factor for the influence of distance at the border when determining initialization or termination hypotheses.", # noqa: E501
+ "Scaling factor for the influence of track-to-track distance on linking probability.", # noqa: E501
+ "Scaling factor for the influence of cell state and position on division (mitosis/branching) probability.", # noqa: E501
+ ]
+
+ scaling_factor_widgets = {}
+ for name, label, tooltip in zip(names, labels, tooltips):
+ widget = QtWidgets.QDoubleSpinBox()
+ widget.setStepType(QtWidgets.QAbstractSpinBox.AdaptiveDecimalStepType)
+ widget.setToolTip(tooltip)
+ scaling_factor_widgets[name] = (label, widget)
+
+ return scaling_factor_widgets
+
+
+def _create_threshold_widgets() -> dict[str, tuple[str, QtWidgets.QWidget]]:
+ """Create widgets for setting thresholds for the HypothesisModel"""
+
+ distance_threshold = QtWidgets.QDoubleSpinBox()
+ distance_threshold.setStepType(QtWidgets.QAbstractSpinBox.AdaptiveDecimalStepType)
+ distance_threshold.setToolTip(
+ "A threshold distance from the edge of the field of view to add an "
+ "initialization or termination hypothesis."
+ )
+ widgets = {"theta_dist": ("distance threshold", distance_threshold)}
+
+ time_threshold = QtWidgets.QDoubleSpinBox()
+ time_threshold.setStepType(QtWidgets.QAbstractSpinBox.AdaptiveDecimalStepType)
+ time_threshold.setToolTip(
+ "A threshold time from the beginning or end of movie to add "
+ "an initialization or termination hypothesis."
+ )
+ widgets["theta_time"] = ("time threshold", time_threshold)
+
+ apoptosis_threshold = QtWidgets.QSpinBox()
+ apoptosis_threshold.setStepType(QtWidgets.QAbstractSpinBox.AdaptiveDecimalStepType)
+ apoptosis_threshold.setToolTip(
+ "Number of apoptotic detections to be considered a genuine event.\n"
+ "Detections are counted consecutively from the back of the track"
+ )
+ widgets["apop_thresh"] = ("apoptosis threshold", apoptosis_threshold)
+
+ return widgets
+
+
+def _create_bin_size_widgets() -> dict[str, tuple[str, QtWidgets.QWidget]]:
+ """Create widget for setting bin sizes for the HypothesisModel"""
+
+ distance_bin_size = QtWidgets.QDoubleSpinBox()
+ distance_bin_size.setStepType(QtWidgets.QAbstractSpinBox.AdaptiveDecimalStepType)
+ distance_bin_size.setToolTip(
+ "Isotropic spatial bin size for considering hypotheses.\n"
+ "Larger bin sizes generate more hypothesese for each tracklet."
+ )
+ widgets = {"dist_thresh": ("distance bin size", distance_bin_size)}
+
+ time_bin_size = QtWidgets.QDoubleSpinBox()
+ time_bin_size.setStepType(QtWidgets.QAbstractSpinBox.AdaptiveDecimalStepType)
+ time_bin_size.setToolTip(
+ "Temporal bin size for considering hypotheses.\n"
+ "Larger bin sizes generate more hypothesese for each tracklet."
+ )
+ widgets["time_thresh"] = ("time bin size", time_bin_size)
+
+ return widgets
+
+
+def create_optimiser_widgets() -> dict[str, tuple[str, QtWidgets.QWidget]]:
+ """Create widgets for setting parameters of the HypothesisModel"""
+
+ widgets = {
+ **_create_hypotheses_widgets(),
+ **_create_scaling_factor_widgets(),
+ **_create_threshold_widgets(),
+ **_create_bin_size_widgets(),
+ }
+
+ segmentation_miss_rate = QtWidgets.QDoubleSpinBox()
+ segmentation_miss_rate.setStepType(
+ QtWidgets.QAbstractSpinBox.AdaptiveDecimalStepType
+ )
+ segmentation_miss_rate.setToolTip(
+ "Miss rate for the segmentation.\n"
+ "e.g. 1/100 segmentations incorrect gives a segmentation miss rate of 0.01."
+ )
+ widgets["segmentation_miss_rate"] = ("miss rate", segmentation_miss_rate)
+
+ relax = QtWidgets.QCheckBox()
+ relax.setToolTip(
+ "Disable the time and distance thresholds.\n"
+ "This means that tracks can initialize or terminate anywhere and"
+ "at any time in the dataset."
+ )
+ relax.setTristate(False) # noqa: FBT003
+ widgets["relax"] = ("relax thresholds", relax)
+
+ return widgets
diff --git a/btrack/napari/widgets/create_ui.py b/btrack/napari/widgets/create_ui.py
index 9cf0533a..ca37703c 100644
--- a/btrack/napari/widgets/create_ui.py
+++ b/btrack/napari/widgets/create_ui.py
@@ -1,32 +1,159 @@
from __future__ import annotations
-from typing import TYPE_CHECKING
+from qtpy import QtWidgets
-if TYPE_CHECKING:
- from magicgui.widgets import Widget
+from napari.viewer import Viewer
from btrack.napari.widgets._general import (
- create_control_widgets,
+ create_basic_widgets,
+ create_config_widgets,
create_input_widgets,
- create_update_method_widgets,
+ create_logo_widgets,
+ create_track_widgets,
)
-from btrack.napari.widgets._hypothesis import create_hypothesis_model_widgets
from btrack.napari.widgets._motion import create_motion_model_widgets
+from btrack.napari.widgets._optimiser import create_optimiser_widgets
-def create_widgets() -> list[Widget]:
- """Create all the widgets for the plugin"""
+class BtrackWidget(QtWidgets.QScrollArea):
+ """Main btrack widget"""
- input_widgets = create_input_widgets()
- update_method_widgets = create_update_method_widgets()
- motion_model_widgets = create_motion_model_widgets()
- hypothesis_model_widgets = create_hypothesis_model_widgets()
- control_buttons = create_control_widgets()
+ def __getitem__(self, key: str) -> QtWidgets.QWidget:
+ return self._widgets[key]
- return [
- *input_widgets,
- *update_method_widgets,
- *motion_model_widgets,
- *hypothesis_model_widgets,
- *control_buttons,
- ]
+ def __init__(self, napari_viewer: Viewer) -> None:
+ """Instantiates the primary widget in napari.
+
+ Args:
+ napari_viewer: A napari viewer instance
+ """
+ super().__init__()
+
+ # We will need to viewer for various callbacks
+ self.viewer = napari_viewer
+
+ # Let the scroll area automatically resize the widget
+ self.setWidgetResizable(True) # noqa: FBT003
+
+ self._main_layout = QtWidgets.QVBoxLayout()
+ self._main_widget = QtWidgets.QWidget()
+ self._main_widget.setLayout(self._main_layout)
+ self.setWidget(self._main_widget)
+ self._tabs = QtWidgets.QTabWidget()
+
+ # Create widgets and add to layout
+ self._widgets = {}
+
+ self._add_logo_widgets()
+ self._add_input_widgets()
+ self._add_track_widgets()
+ # This must be added after the track widget
+ self._main_layout.addWidget(self._tabs, stretch=0)
+ self._add_basic_widgets()
+ self._add_motion_model_widgets()
+ self._add_optimiser_widgets()
+ self._add_config_widgets()
+
+ # Expand the main widget
+ self._main_layout.addStretch(stretch=1)
+
+ # Add attribute access for each widget
+ for name, widget in self._widgets.items():
+ self.__setattr__(
+ name,
+ widget,
+ )
+
+ def _add_logo_widgets(self) -> None:
+ """Adds the btrack logo with a link to the documentation"""
+ logo_widgets = create_logo_widgets()
+ self._widgets.update(logo_widgets)
+ for widget in logo_widgets.values():
+ self._main_layout.addWidget(widget, stretch=0)
+
+ def _add_input_widgets(self) -> None:
+ """Create input widgets and add to main layout"""
+ labels_and_widgets = create_input_widgets()
+ self._widgets.update(
+ {key: value[1] for key, value in labels_and_widgets.items()}
+ )
+
+ widget_holder = QtWidgets.QGroupBox("Input")
+ layout = QtWidgets.QFormLayout()
+ for label, widget in labels_and_widgets.values():
+ label_widget = QtWidgets.QLabel(label)
+ label_widget.setToolTip(widget.toolTip())
+ layout.addRow(label_widget, widget)
+ widget_holder.setLayout(layout)
+ self._main_layout.addWidget(widget_holder, stretch=0)
+
+ def _add_basic_widgets(self) -> None:
+ """Create update method widgets and add to main layout"""
+ labels_and_widgets = create_basic_widgets()
+ self._widgets.update(
+ {key: value[1] for key, value in labels_and_widgets.items()}
+ )
+
+ layout = QtWidgets.QFormLayout()
+ for label, widget in labels_and_widgets.values():
+ label_widget = QtWidgets.QLabel(label)
+ label_widget.setToolTip(widget.toolTip())
+ layout.addRow(label_widget, widget)
+
+ tab = QtWidgets.QWidget()
+ tab.setLayout(layout)
+ self._tabs.addTab(tab, "Basic")
+
+ def _add_motion_model_widgets(self) -> None:
+ """Create motion model widgets and add to main layout"""
+ labels_and_widgets = create_motion_model_widgets()
+ self._widgets.update(
+ {key: value[1] for key, value in labels_and_widgets.items()}
+ )
+
+ layout = QtWidgets.QFormLayout()
+ for label, widget in labels_and_widgets.values():
+ label_widget = QtWidgets.QLabel(label)
+ label_widget.setToolTip(widget.toolTip())
+ layout.addRow(label_widget, widget)
+
+ tab = QtWidgets.QWidget()
+ tab.setLayout(layout)
+ self._tabs.addTab(tab, "Motion")
+
+ def _add_optimiser_widgets(self) -> None:
+ """Create hypothesis model widgets and add to main layout"""
+ labels_and_widgets = create_optimiser_widgets()
+ self._widgets.update(
+ {key: value[1] for key, value in labels_and_widgets.items()}
+ )
+
+ layout = QtWidgets.QFormLayout()
+ for label, widget in labels_and_widgets.values():
+ label_widget = QtWidgets.QLabel(label)
+ label_widget.setToolTip(widget.toolTip())
+ layout.addRow(label_widget, widget)
+
+ tab = QtWidgets.QWidget()
+ tab.setLayout(layout)
+ self._tabs.addTab(tab, "Optimiser")
+
+ def _add_config_widgets(self) -> None:
+ """Creates the IO widgets related to the user config"""
+ io_widgets = create_config_widgets()
+ self._widgets.update(io_widgets)
+
+ layout = QtWidgets.QFormLayout()
+ for widget in io_widgets.values():
+ layout.addRow(widget)
+
+ tab = QtWidgets.QWidget()
+ tab.setLayout(layout)
+ self._tabs.addTab(tab, "Config")
+
+ def _add_track_widgets(self) -> None:
+ """Create widgets for running the tracking"""
+ track_widgets = create_track_widgets()
+ self._widgets.update(track_widgets)
+ for widget in track_widgets.values():
+ self._main_layout.addWidget(widget, stretch=0)
diff --git a/btrack/napari/writer.py b/btrack/napari/writer.py
new file mode 100644
index 00000000..5ea73de6
--- /dev/null
+++ b/btrack/napari/writer.py
@@ -0,0 +1,30 @@
+"""
+This module is a writer plugin to export Tracks layers using BTrack
+"""
+from typing import Optional
+
+import numpy.typing as npt
+
+from btrack.io import HDF5FileHandler
+from btrack.utils import napari_to_tracks
+
+
+def export_to_hdf(
+ path: str,
+ data: npt.ArrayLike,
+ meta: dict,
+) -> Optional[str]:
+ tracks = napari_to_tracks(
+ data=data,
+ properties=meta.get("properties", {}),
+ graph=meta.get("graph", {}),
+ )
+
+ with HDF5FileHandler(
+ filename=path,
+ read_write="w",
+ obj_type="obj_type_1",
+ ) as writer:
+ writer.write_tracks(tracks)
+
+ return path
diff --git a/btrack/optimise/hypothesis.py b/btrack/optimise/hypothesis.py
index 14404568..f55765e6 100644
--- a/btrack/optimise/hypothesis.py
+++ b/btrack/optimise/hypothesis.py
@@ -18,6 +18,7 @@
import ctypes
+from typing import ClassVar
from btrack import constants
@@ -49,7 +50,7 @@ class Hypothesis(ctypes.Structure):
These are automatically generated by the optimiser
"""
- _fields_ = [
+ _fields_: ClassVar[list] = [
("hypothesis", ctypes.c_uint),
("ID", ctypes.c_uint),
("probability", ctypes.c_double),
@@ -94,7 +95,7 @@ class PyHypothesisParams(ctypes.Structure):
"""
- _fields_ = [
+ _fields_: ClassVar[list] = [
("lambda_time", ctypes.c_double),
("lambda_dist", ctypes.c_double),
("lambda_link", ctypes.c_double),
diff --git a/btrack/optimise/optimiser.py b/btrack/optimise/optimiser.py
index 9a5dbdb9..a002ba06 100644
--- a/btrack/optimise/optimiser.py
+++ b/btrack/optimise/optimiser.py
@@ -17,7 +17,6 @@
__email__ = "a.lowe@ucl.ac.uk"
import logging
-from typing import List
from cvxopt import matrix, spmatrix
from cvxopt.glpk import ilp
@@ -101,7 +100,7 @@ class TrackOptimiser:
"""
def __init__(self, options: dict = GLPK_OPTIONS):
- self._hypotheses: List[hypothesis.Hypothesis] = []
+ self._hypotheses: list[hypothesis.Hypothesis] = []
self.options = options # TODO(arl): do some option parsing?
@property
diff --git a/btrack/src/tracker.cc b/btrack/src/tracker.cc
index 14ffc172..51ae65c8 100644
--- a/btrack/src/tracker.cc
+++ b/btrack/src/tracker.cc
@@ -19,14 +19,14 @@
using namespace ProbabilityDensityFunctions;
using namespace BayesianUpdateFunctions;
-void write_belief_matrix_to_CSV(std::string a_filename,
- Eigen::Ref a_belief) {
- std::cout << "Writing: " << a_filename << std::endl;
- std::ofstream belief_file;
- belief_file.open(a_filename);
- belief_file << a_belief.format(CSVFormat);
- belief_file.close();
-}
+// void write_belief_matrix_to_CSV(std::string a_filename,
+// Eigen::Ref a_belief) {
+// std::cout << "Writing: " << a_filename << std::endl;
+// std::ofstream belief_file;
+// belief_file.open(a_filename);
+// belief_file << a_belief.format(CSVFormat);
+// belief_file.close();
+// }
// set up the tracker using an existing track manager
BayesianTracker::BayesianTracker(const bool verbose,
@@ -52,12 +52,12 @@ BayesianTracker::BayesianTracker(const bool verbose,
// set the appropriate cost function
set_update_mode(update_mode);
- // set a outputfile path
- // define a filepath for debugging output
- if (WRITE_BELIEF_MATRIX) {
- m_debug_filepath = std::filesystem::temp_directory_path();
- std::cout << "Using temp file path: " << m_debug_filepath << std::endl;
- }
+ // // set a outputfile path
+ // // define a filepath for debugging output
+ // if (WRITE_BELIEF_MATRIX) {
+ // m_debug_filepath = std::experimental::filesystem::temp_directory_path();
+ // std::cout << "Using temp file path: " << m_debug_filepath << std::endl;
+ // }
}
BayesianTracker::~BayesianTracker() {
@@ -340,13 +340,13 @@ void BayesianTracker::step(const unsigned int steps) {
update_iteration++;
}
- // write out belief matrix here
- if (WRITE_BELIEF_MATRIX) {
- std::stringstream belief_filename;
- belief_filename << m_debug_filepath << "belief_" << current_frame
- << ".csv";
- write_belief_matrix_to_CSV(belief_filename.str(), belief);
- }
+ // // write out belief matrix here
+ // if (WRITE_BELIEF_MATRIX) {
+ // std::stringstream belief_filename;
+ // belief_filename << m_debug_filepath << "belief_" << current_frame
+ // << ".csv";
+ // write_belief_matrix_to_CSV(belief_filename.str(), belief);
+ // }
// if we're storing the graph edges for future optimization, do so here
// this should be done *BEFORE* linking because it relies on the
diff --git a/btrack/utils.py b/btrack/utils.py
index ca4f03b1..a4be10a0 100644
--- a/btrack/utils.py
+++ b/btrack/utils.py
@@ -3,14 +3,19 @@
import dataclasses
import functools
import logging
-from typing import List, Optional
+from typing import TYPE_CHECKING, Optional
+
+if TYPE_CHECKING:
+ import numpy.typing as npt
import numpy as np
from skimage.util import map_array
# import core
from . import _version, btypes, constants
+from .btypes import Tracklet
from .constants import DEFAULT_EXPORT_PROPERTIES, Dimensionality
+from .io import objects_from_dict
from .io._localization import segmentation_to_objects
from .models import HypothesisModel, MotionModel, ObjectModel
@@ -27,10 +32,7 @@
def log_error(err_code) -> bool:
"""Take an error code from the tracker and log an error for the user."""
error = constants.Errors(err_code)
- if (
- error != constants.Errors.SUCCESS
- and error != constants.Errors.NO_ERROR
- ):
+ if error not in [constants.Errors.SUCCESS, constants.Errors.NO_ERROR]:
logger.error(f"ERROR: {error}")
return True
return False
@@ -65,23 +67,17 @@ def log_stats(stats: dict) -> None:
def read_motion_model(cfg: dict) -> Optional[MotionModel]:
cfg = cfg.get("MotionModel", {})
- if not cfg:
- return None
- return MotionModel(**cfg)
+ return MotionModel(**cfg) if cfg else None
def read_object_model(cfg: dict) -> Optional[ObjectModel]:
cfg = cfg.get("ObjectModel", {})
- if not cfg:
- return None
- return ObjectModel(**cfg)
+ return ObjectModel(**cfg) if cfg else None
def read_hypothesis_model(cfg: dict) -> Optional[HypothesisModel]:
cfg = cfg.get("HypothesisModel", {})
- if not cfg:
- return None
- return HypothesisModel(**cfg)
+ return HypothesisModel(**cfg) if cfg else None
def crop_volume(objects, volume=constants.VOLUME):
@@ -89,21 +85,17 @@ def crop_volume(objects, volume=constants.VOLUME):
axes = zip(["x", "y", "z", "t"], volume)
def within(o):
- return all(
- getattr(o, a) >= v[0] and getattr(o, a) <= v[1] for a, v in axes
- )
+ return all(getattr(o, a) >= v[0] and getattr(o, a) <= v[1] for a, v in axes)
return [o for o in objects if within(o)]
-def _lbep_table(tracks: List[btypes.Tracklet]) -> np.array:
+def _lbep_table(tracks: list[btypes.Tracklet]) -> npt.NDArray:
"""Create an LBEP table from a track."""
return np.asarray([trk.LBEP() for trk in tracks], dtype=np.int32)
-def _cat_tracks_as_dict(
- tracks: list[btypes.Tracklet], properties: List[str]
-) -> dict:
+def _cat_tracks_as_dict(tracks: list[btypes.Tracklet], properties: list[str]) -> dict:
"""Concatenate all tracks as dictionary."""
assert all(isinstance(t, btypes.Tracklet) for t in tracks)
@@ -188,17 +180,19 @@ def tracks_to_napari(
with dimensions (5,) would be split into `softmax-0` ... `softmax-4` for
representation in napari.
"""
- # guess the dimensionality from the data by checking whether the z values
+ # guess the dimensionality from the data by checking whether the non-dummy z values
# are all zero. If all z are zero then the data are planar, i.e. 2D
if ndim is None:
- z = np.concatenate([track.z for track in tracks])
+ z = np.concatenate(
+ [np.asarray(track.z)[~np.asarray(track.dummy)] for track in tracks]
+ )
ndim = Dimensionality.THREE if np.any(z) else Dimensionality.TWO
if ndim not in (Dimensionality.TWO, Dimensionality.THREE):
raise ValueError("ndim must be 2 or 3 dimensional.")
t_header = ["ID", "t"] + ["z", "y", "x"][-ndim:]
- p_header = ["t", "state", "generation", "root", "parent"]
+ p_header = ["t", "state", "generation", "root", "parent", "dummy"]
# ensure lexicographic ordering of tracks
ordered = sorted(tracks, key=lambda t: t.ID)
@@ -209,9 +203,7 @@ def tracks_to_napari(
prop_keys = p_header + [k for k in tracks_as_dict if k not in t_header]
# get the data for napari
- data = np.stack(
- [v for k, v in tracks_as_dict.items() if k in t_header], axis=1
- )
+ data = np.stack([v for k, v in tracks_as_dict.items() if k in t_header], axis=1)
properties = {k: v for k, v in tracks_as_dict.items() if k in prop_keys}
# replace any NaNs in the properties with an interpolated value
@@ -228,27 +220,114 @@ def nans_idx(x):
return data, properties, graph
+def napari_to_tracks(
+ data: npt.NDArray,
+ properties: Optional[dict[str, npt.ArrayLike]],
+ graph: Optional[dict[int, list[int]]],
+) -> list[btypes.Tracklet]:
+ """Convert napari Tracks to a list of Tracklets.
+
+ Parameters
+ ----------
+ data : array (N, D+1)
+ Coordinates for N points in D+1 dimensions. ID,T,(Z),Y,X. The first
+ axis is the integer ID of the track. D is either 3 or 4 for planar
+ or volumetric timeseries respectively.
+ properties : dict {str: array (N,)}
+ Properties for each point. Each property should be an array of length N,
+ where N is the number of points.
+ graph : dict {int: list}
+ Graph representing associations between tracks. Dictionary defines the
+ mapping between a track ID and the parents of the track. This can be
+ one (the track has one parent, and the parent has >=1 child) in the
+ case of track splitting, or more than one (the track has multiple
+ parents, but only one child) in the case of track merging.
+
+ Returns
+ -------
+ tracks : list[btypes.Tracklet]
+ A list of tracklet objects created from the napari Tracks layer data.
+
+ """
+
+ if data.shape[1] == Dimensionality.FIVE:
+ track_id, t, z, y, x = data.T
+ elif data.shape[1] == Dimensionality.FOUR:
+ track_id, t, y, x = data.T
+ z = np.zeros_like(x)
+ else:
+ raise ValueError(
+ "Data must have either 4 (ID, t, y, x) or 5 (ID, t, z, y, x) columns, "
+ f"not {data.shape[1]}"
+ )
+
+ # Create all PyTrackObjects
+ objects_dict = {
+ "ID": np.arange(track_id.size),
+ "t": t,
+ "x": x,
+ "y": y,
+ "z": z,
+ "dummy": properties.get("dummy", np.full_like(track_id, fill_value=False)),
+ "label": properties.get(
+ "state", np.full_like(track_id, fill_value=constants.States.NULL)
+ ),
+ }
+ track_objects = objects_from_dict(objects_dict)
+
+ # Create all Tracklets
+ tracklets = []
+ for track in np.unique(track_id).astype(int):
+ # Create tracklet
+ track_indices = np.argwhere(track_id == track).ravel()
+ track_data = [track_objects[i] for i in track_indices]
+ parent = graph.get(track, [track])[0]
+ children = [child for (child, parents) in graph.items() if track in parents]
+ tracklet = Tracklet(
+ ID=track,
+ data=track_data,
+ parent=parent,
+ children=children,
+ )
+
+ # Determine root tracklet
+ tracklet.root = parent
+ tracklet.generation = 0 if tracklet.root == track else 1
+ while tracklet.root in graph:
+ tracklet.root = graph[tracklet.root][0]
+ tracklet.generation += 1
+
+ tracklets.append(tracklet)
+
+ return tracklets
+
+
def update_segmentation(
- segmentation: np.ndarray,
+ segmentation: npt.NDArray,
tracks: list[btypes.Tracklet],
*,
+ scale: Optional[tuple(float)] = None,
color_by: str = "ID",
-) -> np.ndarray:
+) -> npt.NDArray:
"""Map tracks back into a masked array.
Parameters
----------
- segmentation : np.array
+ segmentation : npt.NDArray
Array containing a timeseries of single cell masks. Dimensions should be
ordered T(Z)YX. Assumes that this is not binary and each object has a unique ID.
tracks : list[btypes.Tracklet]
A list of :py:class:`btrack.btypes.Tracklet` objects from BayesianTracker.
+ scale : tuple, optional
+ A scale for each spatial dimension of the input tracks. Defaults
+ to one for all axes, and allows scaling for anisotropic imaging data.
+ Dimensions should be ordered XY(Z).
color_by : str, default = "ID"
A value to recolor the segmentation by.
Returns
-------
- relabeled : np.array
+ relabeled : npt.NDArray
Array containing the same masks as segmentation but relabeled to
maintain single cell identity over time.
@@ -267,13 +346,26 @@ def update_segmentation(
keys = {k: i for i, k in enumerate(DEFAULT_EXPORT_PROPERTIES)}
+ keys.update(
+ {
+ key: idx
+ for idx, key in enumerate(
+ tracks[0].properties.keys(), start=max(keys.values()) + 1
+ )
+ }
+ )
+
coords_arr = np.concatenate(
- [
- track.to_array()[~np.array(track.dummy), : len(keys)].astype(int)
- for track in tracks
- ]
+ [track.to_array()[~np.array(track.dummy), :].astype(int) for track in tracks]
)
+ scale = tuple([1.0] * (segmentation.ndim - 1)) if scale is None else scale
+
+ if (segmentation.ndim - 1) != len(scale):
+ raise ValueError(
+ "Scale should have the same number of spatial dimensions as `segmentation`."
+ )
+
if color_by not in keys:
raise ValueError(f"Property ``{color_by}`` not found in track.")
@@ -284,10 +376,14 @@ def update_segmentation(
xc, yc = frame_coords[:, keys["x"]], frame_coords[:, keys["y"]]
new_id = frame_coords[:, keys[color_by]]
+ xc = (xc * scale[0]).astype(int)
+ yc = (yc * scale[1]).astype(int)
+
if single_segmentation.ndim == constants.Dimensionality.TWO:
old_id = single_segmentation[yc, xc]
elif single_segmentation.ndim == constants.Dimensionality.THREE:
zc = frame_coords[:, keys["z"]]
+ zc = (zc * scale[2]).astype(int)
old_id = single_segmentation[zc, yc, xc]
relabeled[t] = map_array(single_segmentation, old_id, new_id) * (
@@ -306,10 +402,7 @@ class SystemInformation:
def __repr__(self) -> str:
# override to have slightly nicer formatting
return "\n".join(
- [
- f"{key}: {value}"
- for key, value in dataclasses.asdict(self).items()
- ]
+ [f"{key}: {value}" for key, value in dataclasses.asdict(self).items()]
)
diff --git a/build.sh b/build.sh
index 5cdd7313..7c983195 100755
--- a/build.sh
+++ b/build.sh
@@ -7,7 +7,7 @@ mkdir ./btrack/obj
# clone Eigen
if [ ! -e ./btrack/include/eigen/signature_of_eigen3_matrix_library ]
then
- git clone https://gitlab.com/libeigen/eigen.git ./btrack/include/eigen
+ git clone --depth 1 --branch 3.3.9 https://gitlab.com/libeigen/eigen.git ./btrack/include/eigen
fi
# build the tracker
diff --git a/codecov.yml b/codecov.yml
index a77aebe7..e5dbe46f 100644
--- a/codecov.yml
+++ b/codecov.yml
@@ -1,5 +1,6 @@
coverage:
status:
+ patch: off
project:
default:
target: auto
diff --git a/docs/dev_guide/index.rst b/docs/dev_guide/index.rst
index 16cbaa2c..f7672fc5 100644
--- a/docs/dev_guide/index.rst
+++ b/docs/dev_guide/index.rst
@@ -15,18 +15,16 @@ If you would rather install the latest development version, and/or compile direc
.. code:: sh
- git clone https://github.com/quantumjot/btrack.git
- conda env create -f ./btrack/environment.yml
- conda activate btrack
- cd btrack
- ./build.sh
- pip install -e .
+ git clone https://github.com/quantumjot/btrack.git
+ cd btrack
+ ./build.sh
+ pip install -e .
If developing the documentation then run the following
.. code:: sh
- pip install -e .[docs]
+ pip install -e .[docs]
Releasing
---------
@@ -35,12 +33,12 @@ Releases are published to PyPI automatically when a tag is pushed to GitHub.
.. code-block:: sh
- # Set next version number
- export RELEASE=x.x.x
+ # Set next version number
+ export RELEASE=x.x.x
- # Create tags
- git commit --allow-empty -m "Release $RELEASE"
- git tag -a v$RELEASE -m "v$RELEASE"
+ # Create tags
+ git commit --allow-empty -m "Release $RELEASE"
+ git tag -a v$RELEASE -m "v$RELEASE"
- # Push
- git push upstream --tags
+ # Push
+ git push upstream --tags
diff --git a/docs/user_guide/configuration.rst b/docs/user_guide/configuration.rst
index 47d7a875..30ba7a42 100644
--- a/docs/user_guide/configuration.rst
+++ b/docs/user_guide/configuration.rst
@@ -20,7 +20,7 @@ The motion model is used to make forward predictions about the location of objec
"measurements": 3,
"states": 6,
"accuracy": 7.5,
- "prob_not_assign": 0.001,
+ "prob_not_assign": 0.1,
"max_lost": 5,
"A": {
"matrix": [1,0,0,1,0,0,
diff --git a/docs/user_guide/installation.rst b/docs/user_guide/installation.rst
index a2c56027..47527f56 100644
--- a/docs/user_guide/installation.rst
+++ b/docs/user_guide/installation.rst
@@ -18,29 +18,29 @@ Installing Scientific Python
If you do have already an Anaconda or miniconda setup you can jump to the next step.
.. note::
- We strongly recommend using a `Python virtual environment `__ or a `conda virtual environment. `__
+ We strongly recommend using a `Python virtual environment `__ or a `conda virtual environment. `__
If you don't currently have a working scientific Python distribution then follow the `Miniconda Python distribution installation instructions `__ to install Miniconda.
.. note::
- Miniconda is a lighter version of conda. But all the commands are the same.
+ Miniconda is a lighter version of conda. But all the commands are the same.
Setting up a conda environment
------------------------------
..
- TODO Set the conda-forge channels
+ TODO Set the conda-forge channels
Once you have ``conda`` installed, you can create a virtual environment from the terminal/system command prompt or the 'Anaconda Prompt' (under Windows) as::
- conda create -n btrack-env
+ conda create -n btrack-env
and access to the environment via::
- conda activate btrack-env
+ conda activate btrack-env
We could have skipped these two steps and install ``btrack`` in the base environment, but virtual environments allow us to keep packages independent of other installations.
@@ -50,6 +50,6 @@ Installing btrack
After we've created and activated the virtual environment, on the same terminal, we install ``btrack`` with::
- pip install btrack
+ pip install btrack
This will download and install ``btrack`` and all its dependencies.
diff --git a/docs/user_guide/napari.rst b/docs/user_guide/napari.rst
index b9f57ec0..45e62ebc 100644
--- a/docs/user_guide/napari.rst
+++ b/docs/user_guide/napari.rst
@@ -9,6 +9,10 @@ dependencies needed to use these plugins, install the ``napari`` extra via.::
pip install btrack[napari]
+If working on Apple Silicon then also run::
+
+ conda install -c conda-forge pyqt
+
The Tracks layer
================
@@ -17,11 +21,11 @@ We developed the ``Tracks`` layer that is now part of the multidimensional image
.. code:: python
- import napari
+ import napari
- viewer = napari.Viewer()
- viewer.add_labels(segmentation)
- viewer.add_tracks(data, properties=properties, graph=graph)
+ viewer = napari.Viewer()
+ viewer.add_labels(segmentation)
+ viewer.add_tracks(data, properties=properties, graph=graph)
Read more about `the tracks API at Napari's documentation `_.
diff --git a/docs/user_guide/saving_tracks.rst b/docs/user_guide/saving_tracks.rst
index d517bbae..575d396a 100644
--- a/docs/user_guide/saving_tracks.rst
+++ b/docs/user_guide/saving_tracks.rst
@@ -19,7 +19,7 @@ Or, if you have a list of :py:class:`btrack.btypes.Tracklet`, you can write them
.. code:: python
- with btrack.dataio.HDF5FileHandler(
+ with btrack.io.HDF5FileHandler(
'/path/to/tracks.h5', 'w', obj_type='obj_type_1'
) as writer:
writer.write_tracks(tracks)
diff --git a/environment.yml b/environment.yml
deleted file mode 100644
index e9ce4555..00000000
--- a/environment.yml
+++ /dev/null
@@ -1,10 +0,0 @@
-name: btrack
-channels:
- - conda-forge
-dependencies:
- - cvxopt>=1.2.0
- - h5py>=2.10.0
- - pip
- - python=3.10
- - pip:
- - -r requirements.txt
diff --git a/models/cell_config.json b/models/cell_config.json
deleted file mode 100644
index 85143a1e..00000000
--- a/models/cell_config.json
+++ /dev/null
@@ -1,68 +0,0 @@
-{
- "TrackerConfig":
- {
- "MotionModel":
- {
- "name": "cell_motion",
- "dt": 1.0,
- "measurements": 3,
- "states": 6,
- "accuracy": 7.5,
- "prob_not_assign": 0.001,
- "max_lost": 5,
- "A": {
- "matrix": [1,0,0,1,0,0,
- 0,1,0,0,1,0,
- 0,0,1,0,0,1,
- 0,0,0,1,0,0,
- 0,0,0,0,1,0,
- 0,0,0,0,0,1]
- },
- "H": {
- "matrix": [1,0,0,0,0,0,
- 0,1,0,0,0,0,
- 0,0,1,0,0,0]
- },
- "P": {
- "sigma": 150.0,
- "matrix": [0.1,0,0,0,0,0,
- 0,0.1,0,0,0,0,
- 0,0,0.1,0,0,0,
- 0,0,0,1,0,0,
- 0,0,0,0,1,0,
- 0,0,0,0,0,1]
- },
- "G": {
- "sigma": 15.0,
- "matrix": [0.5,0.5,0.5,1,1,1]
-
- },
- "R": {
- "sigma": 5.0,
- "matrix": [1,0,0,
- 0,1,0,
- 0,0,1]
- }
- },
- "ObjectModel":
- {},
- "HypothesisModel":
- {
- "name": "cell_hypothesis",
- "hypotheses": ["P_FP", "P_init", "P_term", "P_link", "P_branch", "P_dead"],
- "lambda_time": 5.0,
- "lambda_dist": 3.0,
- "lambda_link": 10.0,
- "lambda_branch": 50.0,
- "eta": 1e-10,
- "theta_dist": 20.0,
- "theta_time": 5.0,
- "dist_thresh": 40,
- "time_thresh": 2,
- "apop_thresh": 5,
- "segmentation_miss_rate": 0.1,
- "apoptosis_rate": 0.001,
- "relax": true
- }
- }
-}
diff --git a/models/cell_config_flat.json b/models/cell_config_flat.json
deleted file mode 100644
index 10cb514b..00000000
--- a/models/cell_config_flat.json
+++ /dev/null
@@ -1,85 +0,0 @@
-{
- "name": "Default",
- "version": "0.5.0",
- "verbose": false,
- "motion_model": {
- "measurements": 3,
- "states": 6,
- "A": [
- 1.0, 0.0, 0.0, 1.0, 0.0, 0.0,
- 0.0, 1.0, 0.0, 0.0, 1.0, 0.0,
- 0.0, 0.0, 1.0, 0.0, 0.0, 1.0,
- 0.0, 0.0, 0.0, 1.0, 0.0, 0.0,
- 0.0, 0.0, 0.0, 0.0, 1.0, 0.0,
- 0.0, 0.0, 0.0, 0.0, 0.0, 1.0
- ],
- "H": [
- 1.0, 0.0, 0.0, 0.0, 0.0, 0.0,
- 0.0, 1.0, 0.0, 0.0, 0.0, 0.0,
- 0.0, 0.0, 1.0, 0.0, 0.0, 0.0
- ],
- "P": [
- 15.0, 0.0, 0.0, 0.0, 0.0, 0.0,
- 0.0, 15.0, 0.0, 0.0, 0.0, 0.0,
- 0.0, 0.0, 15.0, 0.0, 0.0, 0.0,
- 0.0, 0.0, 0.0, 150.0, 0.0, 0.0,
- 0.0, 0.0, 0.0, 0.0, 150.0, 0.0,
- 0.0, 0.0, 0.0, 0.0, 0.0, 150.0
- ],
- "R": [
- 5.0, 0.0, 0.0,
- 0.0, 5.0, 0.0,
- 0.0, 0.0, 5.0
- ],
- "G": [ 7.5, 7.5, 7.5, 15.0, 15.0, 15.0],
- "Q": [
- 56.25, 56.25, 56.25, 112.5, 112.5, 112.5,
- 56.25, 56.25, 56.25, 112.5, 112.5, 112.5,
- 56.25, 56.25, 56.25, 112.5, 112.5, 112.5,
- 112.5, 112.5, 112.5, 225.0, 225.0, 225.0,
- 112.5, 112.5, 112.5, 225.0, 225.0, 225.0,
- 112.5, 112.5, 112.5, 225.0, 225.0, 225.0
- ],
- "dt": 1.0,
- "accuracy": 7.5,
- "max_lost": 5,
- "prob_not_assign": 0.001,
- "name": "cell_motion"
- },
- "object_model": null,
- "hypothesis_model": {
- "hypotheses": [
- "P_FP",
- "P_init",
- "P_term",
- "P_link",
- "P_branch",
- "P_dead"
- ],
- "lambda_time": 5.0,
- "lambda_dist": 3.0,
- "lambda_link": 10.0,
- "lambda_branch": 50.0,
- "eta": 1e-10,
- "theta_dist": 20.0,
- "theta_time": 5.0,
- "dist_thresh": 37.0,
- "time_thresh": 2.0,
- "apop_thresh": 5,
- "segmentation_miss_rate": 0.1,
- "apoptosis_rate": 0.001,
- "relax": true,
- "name": "cell_hypothesis"
- },
- "max_search_radius": 100.0,
- "return_kalman": false,
- "volume": null,
- "update_method": 0,
- "optimizer_options": {
- "tm_lim": 60000
- },
- "features": [],
- "tracking_updates": [
- 1
- ]
-}
diff --git a/models/particle_config.json b/models/particle_config.json
deleted file mode 100644
index c7cb999b..00000000
--- a/models/particle_config.json
+++ /dev/null
@@ -1,68 +0,0 @@
-{
- "TrackerConfig":
- {
- "MotionModel":
- {
- "name": "particle_motion",
- "dt": 1.0,
- "measurements": 3,
- "states": 6,
- "accuracy": 7.5,
- "prob_not_assign": 0.001,
- "max_lost": 5,
- "A": {
- "matrix": [1,0,0,0,0,0,
- 0,1,0,0,0,0,
- 0,0,1,0,0,0,
- 0,0,0,1,0,0,
- 0,0,0,0,1,0,
- 0,0,0,0,0,1]
- },
- "H": {
- "matrix": [1,0,0,0,0,0,
- 0,1,0,0,0,0,
- 0,0,1,0,0,0]
- },
- "P": {
- "sigma": 150.0,
- "matrix": [0.1,0,0,0,0,0,
- 0,0.1,0,0,0,0,
- 0,0,0.1,0,0,0,
- 0,0,0,1,0,0,
- 0,0,0,0,1,0,
- 0,0,0,0,0,1]
- },
- "G": {
- "sigma": 15.0,
- "matrix": [0.5,0.5,0.5,1,1,1]
-
- },
- "R": {
- "sigma": 5.0,
- "matrix": [1,0,0,
- 0,1,0,
- 0,0,1]
- }
- },
- "ObjectModel":
- {},
- "HypothesisModel":
- {
- "name": "particle_hypothesis",
- "hypotheses": ["P_FP", "P_init", "P_term", "P_link"],
- "lambda_time": 5.0,
- "lambda_dist": 3.0,
- "lambda_link": 10.0,
- "lambda_branch": 50.0,
- "eta": 1e-10,
- "theta_dist": 20.0,
- "theta_time": 5.0,
- "dist_thresh": 40,
- "time_thresh": 2,
- "apop_thresh": 5,
- "segmentation_miss_rate": 0.1,
- "apoptosis_rate": 0.001,
- "relax": true
- }
- }
-}
diff --git a/pyproject.toml b/pyproject.toml
index 5b096a94..81c10341 100644
--- a/pyproject.toml
+++ b/pyproject.toml
@@ -3,7 +3,6 @@ build-backend = "setuptools.build_meta"
requires = [
"setuptools",
"setuptools-scm",
- "wheel",
]
[project]
@@ -18,20 +17,34 @@ classifiers = [
"Programming Language :: C++",
"Programming Language :: Python :: 3",
"Programming Language :: Python :: 3 :: Only",
- "Programming Language :: Python :: 3.8",
"Programming Language :: Python :: 3.9",
"Programming Language :: Python :: 3.10",
+ "Programming Language :: Python :: 3.11",
"Topic :: Scientific/Engineering :: Bio-Informatics",
"Topic :: Scientific/Engineering :: Image Recognition",
"Topic :: Scientific/Engineering :: Visualization",
]
+dependencies = [
+ "cvxopt>=1.3.1",
+ "h5py>=2.10.0",
+ "numpy>=1.17.3",
+ "pandas>=2.0.3",
+ "pooch>=1.0.0",
+ "pydantic<2",
+ "scikit-image>=0.16.2",
+ "scipy>=1.3.1",
+ "tqdm>=4.65.0",
+]
description = "A framework for Bayesian multi-object tracking"
dynamic = [
- "dependencies",
"version",
]
name = "btrack"
-optional-dependencies = {docs = [
+optional-dependencies = {"dev" = [
+ "black",
+ "pre-commit",
+ "ruff",
+], docs = [
"numpydoc",
"pytz",
"sphinx",
@@ -43,18 +56,9 @@ optional-dependencies = {docs = [
"napari-plugin-engine>=0.1.4",
"napari>=0.4.16",
"qtpy",
-], pyqt = [
- "PyQt5 >= 5.12.3, != 5.15.0",
- "btrack[napari]",
-], pyside = [
- "PySide2 >= 5.13.2, != 5.15.0 ; python_version != '3.8'",
- "PySide2 >= 5.14.2, != 5.15.0 ; python_version == '3.8'",
- "btrack[napari]",
-], qt = [
- "btrack[pyside]",
]}
readme = "README.md"
-requires-python = ">=3.8"
+requires-python = ">=3.9"
entry-points."napari.manifest".btrack = "btrack:napari.yaml"
license.file = "LICENSE.md"
urls.bugtracker = "https://github.com/quantumjot/btrack/issues"
@@ -62,30 +66,6 @@ urls.documentation = "https://btrack.readthedocs.io"
urls.homepage = "https://github.com/quantumjot/btrack"
urls.usersupport = "https://github.com/quantumjot/btrack/discussions"
-[tool.black]
-exclude = '''
-(
- /(
- \.eggs
- | \.git
- | \.hg
- | \.mypy_cache
- | \.tox
- | \.venv
- | _build
- | build
- | dist
- | examples
- )/
-)
-'''
-line-length = 79
-target-version = [
- "py38",
- "py39",
- "py310",
-]
-
[tool.coverage]
report = {skip_covered = true, sort = "cover"}
run = {branch = true, parallel = true, source = [
@@ -149,7 +129,7 @@ select = [
"W",
"YTT",
]
-target-version = "py38"
+target-version = "py39"
isort.known-first-party = [
"black",
]
@@ -173,7 +153,6 @@ pep8-naming.classmethod-decorators = [
]
[tool.setuptools]
-dynamic.dependencies.file = "requirements.txt"
packages.find = {}
[tool.setuptools_scm]
@@ -191,38 +170,34 @@ overrides."tool.ruff.isort.section-order".inline_arrays = false
[tool.tox]
legacy_tox_ini = """
-[gh-actions]
-python =
- 3.8: py38
- 3.9: py39
- 3.10: py310
+ [gh-actions]
+ python =
+ 3.9: py39
+ 3.10: py310
+ 3.11: py311
+
+ [gh-actions:env]
+ OS =
+ ubuntu-latest: linux
+ macos-latest: macos
+ windows-latest: windows
-[testenv]
-deps =
- dask
- magicgui
- napari
- pyqt5
- pytest
- pytest-cov
- pytest-qt
- pytest-xvfb ; sys_platform == 'linux'
- qtpy
-conda_deps =
- # Use conda to install cvxopt so tests work on macOS
- cvxopt
-conda_channels = conda-forge
-commands = pytest --cov --cov-report=xml
-passenv =
- CI
- DISPLAY
- GITHUB_ACTIONS
- NUMPY_EXPERIMENTAL_ARRAY_FUNCTION
- PYVISTA_OFF_SCREEN
- XAUTHORITY
+ [testenv]
+ commands = pytest --cov --cov-report=xml
+ deps =
+ napari
+ pyqt5
+ pytest-cov
+ pytest-qt
+ pytest-xvfb ; sys_platform == 'linux'
+ passenv =
+ CI
+ DISPLAY
+ GITHUB_ACTIONS
+ NUMPY_EXPERIMENTAL_ARRAY_FUNCTION
+ PYVISTA_OFF_SCREEN
+ XAUTHORITY
-[tox]
-envlist = py{38,39,310}
-isolated_build = True
-requires = tox-conda
+ [tox]
+ envlist = py{39,310,311}-{linux,macos,windows}
"""
diff --git a/requirements.txt b/requirements.txt
deleted file mode 100644
index 38e2206b..00000000
--- a/requirements.txt
+++ /dev/null
@@ -1,8 +0,0 @@
-cvxopt>=1.2.0
-h5py>=2.10.0
-numpy>=1.17.3
-pooch>=1.0.0
-pydantic>=1.9.0
-scikit-image>=0.16.2
-scipy>=1.3.1
-tqdm
diff --git a/tests/_utils.py b/tests/_utils.py
index 52c379b5..e11879af 100644
--- a/tests/_utils.py
+++ b/tests/_utils.py
@@ -1,14 +1,14 @@
from pathlib import Path
-from typing import Any, Dict, List, Optional, Tuple
+from typing import Any, Optional
import numpy as np
+from numpy import typing as npt
from skimage.measure import label
import btrack
+import btrack.datasets
-CONFIG_FILE = (
- Path(__file__).resolve().parent.parent / "models" / "cell_config.json"
-)
+CONFIG_FILE = btrack.datasets.cell_config()
TEST_DATA_PATH = Path(__file__).resolve().parent / "_test_data"
@@ -18,7 +18,7 @@
def create_test_object(
test_id: Optional[int] = None,
ndim: int = 3,
-) -> Tuple[btrack.btypes.PyTrackObject, Dict[str, Any]]:
+) -> tuple[btrack.btypes.PyTrackObject, dict[str, Any]]:
"""Create a test object."""
rng = np.random.default_rng(seed=RANDOM_SEED)
@@ -39,28 +39,22 @@ def create_test_object(
return obj, data
-def create_test_properties() -> Dict[str, float]:
+def create_test_properties() -> dict:
"""Create test properties for an object."""
rng = np.random.default_rng(seed=RANDOM_SEED)
- properties = {
+ return {
"speed": rng.uniform(0.0, 1.0),
"circularity": rng.uniform(0.0, 1.0),
"reporter": rng.uniform(0.0, 1.0),
"nD": rng.uniform(0.0, 1.0, size=(5,)),
}
- return properties
def create_test_tracklet(
track_len: int,
track_id: Optional[int] = None,
ndim: int = 3,
-) -> Tuple[
- btrack.btypes.Tracklet,
- List[btrack.btypes.PyTrackObject],
- List[Dict[str, Any]],
- int,
-]:
+) -> tuple[btrack.btypes.Tracklet, list[btrack.btypes.PyTrackObject], dict, int]:
"""Create a test track."""
rng = np.random.default_rng(seed=RANDOM_SEED)
@@ -74,9 +68,7 @@ def create_test_tracklet(
tracklet.root = track_id
# convert to dictionary {key: [p0,...,pn]}
- properties = (
- {} if not props else {k: [p[k] for p in props] for k in props[0]}
- )
+ properties = {k: [p[k] for p in props] for k in props[0]} if props else {}
return tracklet, data, properties, track_id
@@ -86,7 +78,7 @@ def create_realistic_tracklet( # noqa: PLR0913
start_y: float,
dx: float,
dy: float,
- track_len: float,
+ track_len: int,
track_ID: int,
) -> btrack.btypes.Tracklet:
"""Create a realistic moving track."""
@@ -95,14 +87,11 @@ def create_realistic_tracklet( # noqa: PLR0913
"x": np.array([start_x + dx * t for t in range(track_len)]),
"y": np.array([start_y + dy * t for t in range(track_len)]),
"t": np.arange(track_len),
- "ID": np.array(
- [(track_ID - 1) * track_len + t for t in range(track_len)]
- ),
+ "ID": np.array([(track_ID - 1) * track_len + t for t in range(track_len)]),
}
objects = btrack.io.objects_from_dict(data)
- track = btrack.btypes.Tracklet(track_ID, objects)
- return track
+ return btrack.btypes.Tracklet(track_ID, objects)
def create_test_image(
@@ -112,7 +101,7 @@ def create_test_image(
binsize: int = 5,
*,
binary: bool = True,
-) -> Tuple[np.ndarray, Optional[np.ndarray]]:
+) -> tuple[npt.NDArray, Optional[npt.NDArray]]:
"""Make a test image that ensures that no two pixels are in contact."""
rng = np.random.default_rng(seed=RANDOM_SEED)
@@ -127,7 +116,7 @@ def create_test_image(
# split this into voxels
bins = boxsize // binsize
- def _sample() -> Tuple[np.ndarray, Tuple[int]]:
+ def _sample() -> tuple[npt.NDArray, tuple]:
_img = np.zeros((binsize,) * ndim, dtype=np.uint16)
_coord = tuple(rng.integers(1, binsize - 1, size=(ndim,)).tolist())
_img[_coord] = 1
@@ -137,9 +126,7 @@ def _sample() -> Tuple[np.ndarray, Tuple[int]]:
return _img, _coord
# now we update nobj grid positions with a sample
- grid = np.stack(np.meshgrid(*[np.arange(bins)] * ndim), -1).reshape(
- -1, ndim
- )
+ grid = np.stack(np.meshgrid(*[np.arange(bins)] * ndim), -1).reshape(-1, ndim)
rbins = rng.choice(grid, size=(nobj,), replace=False)
@@ -147,9 +134,7 @@ def _sample() -> Tuple[np.ndarray, Tuple[int]]:
centroids = []
for v, bin in enumerate(rbins): # noqa: A001
sample, point = _sample()
- slices = tuple(
- [slice(b * binsize, b * binsize + binsize, 1) for b in bin]
- )
+ slices = tuple(slice(b * binsize, b * binsize + binsize, 1) for b in bin)
val = 1 if binary else v + 1
img[slices] = sample * val
@@ -168,9 +153,7 @@ def _sample() -> Tuple[np.ndarray, Tuple[int]]:
), "Number of created centroids != requested in test image."
vals = np.unique(img)
- assert (
- np.max(vals) == 1 if binary else nobj
- ), "Test image labels are incorrect."
+ assert np.max(vals) == 1 if binary else nobj, "Test image labels are incorrect."
return img, centroids_sorted
@@ -181,7 +164,7 @@ def create_test_segmentation_and_tracks(
ndim: int = 2,
*,
binary: bool = False,
-) -> Tuple[np.ndarray, np.ndarray, List[btrack.btypes.Tracklet]]:
+) -> tuple[npt.NDArray, npt.NDArray, list[btrack.btypes.Tracklet]]:
"""Create a test segmentation with four tracks."""
if ndim not in (btrack.constants.Dimensionality.TWO,):
@@ -200,20 +183,14 @@ def create_test_segmentation_and_tracks(
track_B = create_realistic_tracklet(
boxsize - padding, boxsize - padding, -dxy, 0, nframes, 2
)
- track_C = create_realistic_tracklet(
- padding, boxsize - padding, 0, -dxy, nframes, 3
- )
- track_D = create_realistic_tracklet(
- boxsize - padding, padding, 0, dxy, nframes, 4
- )
+ track_C = create_realistic_tracklet(padding, boxsize - padding, 0, -dxy, nframes, 3)
+ track_D = create_realistic_tracklet(boxsize - padding, padding, 0, dxy, nframes, 4)
tracks = [track_A, track_B, track_C, track_D]
# set the segmentation values
for track in tracks:
- t, y, x = np.split(
- track.to_array(properties=["t", "y", "x"]).astype(int), 3, 1
- )
+ t, y, x = np.split(track.to_array(properties=["t", "y", "x"]).astype(int), 3, 1)
segmentation[t, y, x] = 1
ground_truth[t, y, x] = track.ID
@@ -225,12 +202,14 @@ def create_test_segmentation_and_tracks(
def full_tracker_example(
- objects: List[btrack.btypes.PyTrackObject], **kwargs
+ objects: list[btrack.btypes.PyTrackObject], **kwargs
) -> btrack.BayesianTracker:
"""Set up a full tracker example. kwargs can supply configuration options."""
# run the tracking
tracker = btrack.BayesianTracker()
- tracker.configure(CONFIG_FILE)
+ cfg = btrack.config.load_config(CONFIG_FILE)
+ cfg.motion_model.prob_not_assign = 0.001
+ tracker.configure(cfg)
for cfg_key, cfg_value in kwargs.items():
setattr(tracker, cfg_key, cfg_value)
tracker.append(objects)
@@ -240,7 +219,7 @@ def full_tracker_example(
return tracker
-def simple_tracker_example() -> Tuple[btrack.BayesianTracker, Dict[str, Any]]:
+def simple_tracker_example() -> tuple[btrack.BayesianTracker, dict[str, Any]]:
"""Run a simple tracker example with some data."""
x = np.array([200, 201, 202, 203, 204, 207, 208])
y = np.array([503, 507, 499, 500, 510, 515, 518])
diff --git a/tests/conftest.py b/tests/conftest.py
index 49782c70..c6c6bad7 100644
--- a/tests/conftest.py
+++ b/tests/conftest.py
@@ -1,8 +1,10 @@
import os
-from typing import List, Union
+from typing import Union
import numpy as np
+import numpy.typing as npt
import pytest
+from qtpy import QtWidgets
import btrack
@@ -14,6 +16,22 @@
)
+def _write_h5_file(file_path: os.PathLike, test_objects) -> os.PathLike:
+ """
+ Write a h5 file with test objects and return path.
+ """
+ with btrack.io.HDF5FileHandler(file_path, "w") as h:
+ h.write_objects(test_objects)
+
+ return file_path
+
+
+@pytest.fixture
+def sample_tracks():
+ """An example tracks dataset"""
+ return btrack.datasets.example_tracks()
+
+
@pytest.fixture
def test_objects():
"""
@@ -31,16 +49,6 @@ def test_real_objects():
return btrack.io.import_CSV(TEST_DATA_PATH / "test_data.csv")
-def write_h5_file(file_path: os.PathLike, test_objects) -> os.PathLike:
- """
- Write a h5 file with test objects and return path.
- """
- with btrack.io.HDF5FileHandler(file_path, "w") as h:
- h.write_objects(test_objects)
-
- return file_path
-
-
@pytest.fixture
def hdf5_file_path(tmp_path, test_objects) -> os.PathLike:
"""
@@ -48,29 +56,27 @@ def hdf5_file_path(tmp_path, test_objects) -> os.PathLike:
Note that this only saves segmentation results, not tracking results.
"""
- return write_h5_file(tmp_path / "test.h5", test_objects)
+ return _write_h5_file(tmp_path / "test.h5", test_objects)
@pytest.fixture(params=["single", "list"])
def hdf5_file_path_or_paths(
tmp_path, test_objects, request
-) -> Union[os.PathLike, List[os.PathLike]]:
+) -> Union[os.PathLike, list[os.PathLike]]:
"""
Create and save a btrack HDF5 file, and return the path.
Note that this only saves segmentation results, not tracking results.
"""
if request.param == "single":
- return write_h5_file(tmp_path / "test.h5", test_objects)
+ return _write_h5_file(tmp_path / "test.h5", test_objects)
elif request.param == "list":
return [
- write_h5_file(tmp_path / "test1.h5", test_objects),
- write_h5_file(tmp_path / "test2.h5", test_objects),
+ _write_h5_file(tmp_path / "test1.h5", test_objects),
+ _write_h5_file(tmp_path / "test2.h5", test_objects),
]
else:
- raise ValueError(
- "Invalid requests.param, must be one of 'single' or 'list'"
- )
+ raise ValueError("Invalid requests.param, must be one of 'single' or 'list'")
@pytest.fixture
@@ -87,3 +93,25 @@ def default_rng():
Create a default PRNG to use for tests.
"""
return np.random.default_rng(seed=RANDOM_SEED)
+
+
+@pytest.fixture
+def track_widget(make_napari_viewer) -> QtWidgets.QWidget:
+ """Provides an instance of the track widget to test"""
+ make_napari_viewer() # make sure there is a viewer available
+ return btrack.napari.main.create_btrack_widget()
+
+
+@pytest.fixture
+def simplistic_tracker_outputs() -> (
+ tuple[npt.NDArray, dict[str, npt.NDArray], dict[int, list]]
+):
+ """Provides simplistic return values of a btrack run.
+ They have the correct types and dimensions, but contain zeros.
+ Useful for mocking the tracker.
+ """
+ n, d = 10, 3
+ data = np.zeros((n, d + 1))
+ properties = {"some_property": np.zeros(n)}
+ graph = {0: [0]}
+ return data, properties, graph
diff --git a/tests/napari/test_dock_widget.py b/tests/napari/test_dock_widget.py
index c301ff48..1a5dc0f0 100644
--- a/tests/napari/test_dock_widget.py
+++ b/tests/napari/test_dock_widget.py
@@ -1,24 +1,16 @@
from __future__ import annotations
-from typing import TYPE_CHECKING
-
-if TYPE_CHECKING:
- from magicgui.widgets import Container
-
import json
from unittest.mock import patch
-import numpy as np
-import numpy.typing as npt
import pytest
import napari
import btrack
+import btrack.datasets
import btrack.napari
import btrack.napari.main
-from btrack import datasets
-from btrack.datasets import cell_config, particle_config
OLD_WIDGET_LAYERS = 1
NEW_WIDGET_LAYERS = 2
@@ -37,14 +29,10 @@ def test_add_widget(make_napari_viewer):
assert len(list(viewer.window._dock_widgets)) == num_dw + 1
-@pytest.fixture
-def track_widget(make_napari_viewer) -> Container:
- """Provides an instance of the track widget to test"""
- make_napari_viewer() # make sure there is a viewer available
- return btrack.napari.main.create_btrack_widget()
-
-
-@pytest.mark.parametrize("config", [cell_config(), particle_config()])
+@pytest.mark.parametrize(
+ "config",
+ [btrack.datasets.cell_config(), btrack.datasets.particle_config()],
+)
def test_config_to_widgets_round_trip(track_widget, config):
"""Tests that going back and forth between
config objects and widgets works as expected.
@@ -53,12 +41,8 @@ def test_config_to_widgets_round_trip(track_widget, config):
expected_config = btrack.config.load_config(config).json()
unscaled_config = btrack.napari.config.UnscaledTrackerConfig(config)
- btrack.napari.sync.update_widgets_from_config(
- unscaled_config, track_widget
- )
- btrack.napari.sync.update_config_from_widgets(
- unscaled_config, track_widget
- )
+ btrack.napari.sync.update_widgets_from_config(unscaled_config, track_widget)
+ btrack.napari.sync.update_config_from_widgets(unscaled_config, track_widget)
actual_config = unscaled_config.scale_config().json()
@@ -66,24 +50,28 @@ def test_config_to_widgets_round_trip(track_widget, config):
assert json.loads(actual_config) == json.loads(expected_config)
-def test_save_button(track_widget):
+@pytest.mark.parametrize("filename", ["user_config"])
+def test_save_button(track_widget, filename):
"""Tests that clicking the save configuration button
triggers a call to btrack.config.save_config with expected arguments.
"""
- unscaled_config = btrack.napari.config.UnscaledTrackerConfig(cell_config())
- unscaled_config.tracker_config.name = (
- "cell" # this is done in in the gui too
+ unscaled_config = btrack.napari.config.UnscaledTrackerConfig(
+ btrack.datasets.cell_config()
)
+ # default config name matches the filename
+ unscaled_config.tracker_config.name = filename
+ unscaled_config.tracker_config.hypothesis_model.name = filename
+ unscaled_config.tracker_config.motion_model.name = filename
expected_config = unscaled_config.scale_config().json()
with patch(
"btrack.napari.widgets.save_path_dialogue_box"
) as save_path_dialogue_box:
- save_path_dialogue_box.return_value = "user_config.json"
- track_widget.save_config_button.clicked()
+ save_path_dialogue_box.return_value = f"{filename}.json"
+ track_widget.save_config_button.click()
- actual_config = btrack.config.load_config("user_config.json").json()
+ actual_config = btrack.config.load_config(f"{filename}.json").json()
# use json.loads to avoid failure in string comparison because e.g "100.0" != "100"
assert json.loads(expected_config) == json.loads(actual_config)
@@ -93,55 +81,45 @@ def test_load_config(track_widget):
"""Tests that another TrackerConfig can be loaded and made the current config."""
# this is set to be 'cell' rather than 'Default'
- original_config_name = track_widget.config.current_choice
+ original_config_name = track_widget.config_name.currentText()
with patch(
"btrack.napari.widgets.load_path_dialogue_box"
) as load_path_dialogue_box:
- load_path_dialogue_box.return_value = cell_config()
- track_widget.load_config_button.clicked()
+ load_path_dialogue_box.return_value = btrack.datasets.cell_config()
+ track_widget.load_config_button.click()
# We didn't override the name, so it should be 'Default'
- new_config_name = track_widget.config.current_choice
+ new_config_name = track_widget.config_name.currentText()
- assert track_widget.config.value == "Default"
+ assert track_widget.config_name.currentText() == "Default"
assert new_config_name != original_config_name
def test_reset_button(track_widget):
"""Tests that clicking the reset button restores the default config values"""
- original_max_search_radius = track_widget.max_search_radius.value
- original_relax = track_widget.relax.value
+ original_max_search_radius = track_widget.max_search_radius.value()
+ original_relax = track_widget.relax.isChecked()
+ original_optimise = track_widget.enable_optimisation.isChecked()
# change some widget values
- track_widget.max_search_radius.value += 10
- track_widget.relax.value = not track_widget.relax
+ track_widget.max_search_radius.setValue(track_widget.max_search_radius.value() + 10)
+ track_widget.relax.setChecked(not track_widget.relax.isChecked())
+ track_widget.enable_optimisation.setChecked(
+ not track_widget.enable_optimisation.isChecked()
+ )
# click reset button - restores defaults of the currently-selected base config
- track_widget.reset_button.clicked()
+ track_widget.reset_button.click()
- new_max_search_radius = track_widget.max_search_radius.value
- new_relax = track_widget.relax.value
+ new_max_search_radius = track_widget.max_search_radius.value()
+ new_relax = track_widget.relax.isChecked()
+ new_optimise = track_widget.enable_optimisation.isChecked()
assert new_max_search_radius == original_max_search_radius
assert new_relax == original_relax
-
-
-@pytest.fixture
-def simplistic_tracker_outputs() -> (
- tuple[npt.NDArray, dict[str, npt.NDArray], dict[int, list]]
-):
- """Provides simplistic return values of a btrack run.
-
- They have the correct types and dimensions, but contain zeros.
- Useful for mocking the tracker.
- """
- n, d = 10, 3
- data = np.zeros((n, d + 1))
- properties = {"some_property": np.zeros(n)}
- graph = {0: [0]}
- return data, properties, graph
+ assert new_optimise == original_optimise
def test_run_button(track_widget, simplistic_tracker_outputs):
@@ -150,10 +128,18 @@ def test_run_button(track_widget, simplistic_tracker_outputs):
"""
with patch("btrack.napari.main._run_tracker") as run_tracker:
run_tracker.return_value = simplistic_tracker_outputs
- segmentation = datasets.example_segmentation()
+ segmentation = btrack.datasets.example_segmentation()
track_widget.viewer.add_labels(segmentation)
+
+ # we need to explicitly add the layer to the ComboBox
+ track_widget.segmentation.setCurrentIndex(0)
+ track_widget.segmentation.setCurrentText(
+ track_widget.viewer.layers[track_widget.segmentation.currentIndex()].name
+ )
+
assert len(track_widget.viewer.layers) == OLD_WIDGET_LAYERS
- track_widget.call_button.clicked()
+ track_widget.track_button.click()
+
assert run_tracker.called
assert len(track_widget.viewer.layers) == NEW_WIDGET_LAYERS
assert isinstance(track_widget.viewer.layers[-1], napari.layers.Tracks)
diff --git a/tests/test_config.py b/tests/test_config.py
index 5ca70fd7..9d48f1a4 100644
--- a/tests/test_config.py
+++ b/tests/test_config.py
@@ -1,5 +1,5 @@
from pathlib import Path
-from typing import Tuple, Union
+from typing import Union
import numpy as np
import pytest
@@ -14,17 +14,15 @@ def _random_config() -> dict:
rng = np.random.default_rng(seed=RANDOM_SEED)
return {
"max_search_radius": rng.uniform(1, 100),
- "update_method": rng.choice(btrack.constants.BayesianUpdates),
+ "update_method": rng.choice(list(btrack.constants.BayesianUpdates)),
"return_kalman": bool(rng.uniform(0, 2)),
"store_candidate_graph": bool(rng.uniform(0, 2)),
"verbose": bool(rng.uniform(0, 2)),
- "volume": tuple([(0, rng.uniform(1, 100)) for _ in range(3)]),
+ "volume": tuple((0, rng.uniform(1, 100)) for _ in range(3)),
}
-def _validate_config(
- cfg: Union[btrack.BayesianTracker, BaseModel], options: dict
-):
+def _validate_config(cfg: Union[btrack.BayesianTracker, BaseModel], options: dict):
for key, value in options.items():
cfg_value = getattr(cfg, key)
# takes care of recursive model definintions (i.e. MotionModel inside
@@ -81,7 +79,7 @@ def test_config_tracker_setters():
_validate_config(tracker.configuration, options)
-def _cfg_dict() -> Tuple[dict, dict]:
+def _cfg_dict() -> tuple[dict, dict]:
cfg_raw = btrack.config.load_config(CONFIG_FILE)
cfg = _random_config()
cfg.update(cfg_raw.dict())
@@ -89,14 +87,14 @@ def _cfg_dict() -> Tuple[dict, dict]:
return cfg, cfg
-def _cfg_file() -> Tuple[Path, dict]:
+def _cfg_file() -> tuple[str, dict]:
filename = CONFIG_FILE
- assert isinstance(filename, Path)
+ assert isinstance(filename, str)
cfg = btrack.config.load_config(filename)
return filename, cfg.dict()
-def _cfg_pydantic() -> Tuple[btrack.config.TrackerConfig, dict]:
+def _cfg_pydantic() -> tuple[btrack.config.TrackerConfig, dict]:
cfg = btrack.config.load_config(CONFIG_FILE)
options = _random_config()
for key, value in options.items():
diff --git a/tests/test_examples.py b/tests/test_examples.py
index f8f4ad97..efcdbfa8 100644
--- a/tests/test_examples.py
+++ b/tests/test_examples.py
@@ -1,4 +1,4 @@
-from btrack import datasets
+import btrack.datasets
def test_pooch_registry():
@@ -7,4 +7,4 @@ def test_pooch_registry():
This will fail if the remote file does not match the hash hard-coded
in btrack.datasets.
"""
- registry_file = datasets._remote_registry() # noqa: F841
+ registry_file = btrack.datasets._remote_registry() # noqa: F841
diff --git a/tests/test_io.py b/tests/test_io.py
index 662d561b..c5ddd147 100644
--- a/tests/test_io.py
+++ b/tests/test_io.py
@@ -56,6 +56,37 @@ def test_hdf5_write_with_properties(hdf5_file_path):
np.testing.assert_allclose(orig.properties[p], read.properties[p])
+@pytest.mark.parametrize("frac_dummies", [0.1, 0.5, 0.9])
+def test_hdf5_write_dummies(hdf5_file_path, test_objects, frac_dummies):
+ """Test writing tracks with a variable proportion of dummy objects."""
+
+ num_dummies = int(len(test_objects) * frac_dummies)
+
+ for obj in test_objects[:num_dummies]:
+ obj.dummy = True
+ obj.ID = -(obj.ID + 1)
+
+ track_id = 1
+ track_with_dummies = btrack.btypes.Tracklet(track_id, test_objects)
+ track_with_dummies.root = track_id
+ track_with_dummies.parent = track_id
+
+ # write them out
+ with btrack.io.HDF5FileHandler(hdf5_file_path, "w") as h:
+ h.write_tracks(
+ [
+ track_with_dummies,
+ ]
+ )
+
+ # read them in
+ with btrack.io.HDF5FileHandler(hdf5_file_path, "r") as h:
+ tracks_from_file = h.tracks
+ objects_from_file = tracks_from_file[0]._data
+
+ assert sum(obj.dummy for obj in objects_from_file) == num_dummies
+
+
@pytest.mark.parametrize("export_format", ["", ".csv", ".h5"])
def test_tracker_export(tmp_path, export_format):
"""Test that file export works using the `export_delegator`."""
@@ -136,3 +167,20 @@ def test_write_hdf_segmentation(hdf5_file_path):
with btrack.io.HDF5FileHandler(hdf5_file_path, "r") as h:
segmentation_from_file = h.segmentation
np.testing.assert_equal(segmentation, segmentation_from_file)
+
+
+def test_hdf_tree(hdf5_file_path, caplog):
+ """Test that the tree function iterates over the files and writes the output
+ to the logger."""
+ n_log_entries = len(caplog.records)
+
+ # first test with an empty tree
+ btrack.io.hdf._h5_tree({})
+
+ assert len(caplog.records) == n_log_entries
+
+ with btrack.io.HDF5FileHandler(hdf5_file_path, "r") as hdf:
+ hdf.tree()
+
+ n_expected_entries = 8
+ assert len(caplog.records) == n_log_entries + n_expected_entries
diff --git a/tests/test_shared_lib.py b/tests/test_shared_lib.py
index 0a6d0bfb..8323efcb 100644
--- a/tests/test_shared_lib.py
+++ b/tests/test_shared_lib.py
@@ -16,7 +16,7 @@ def test_load_library():
def test_fails_load_library_debug(tmp_path):
"""Test loading a fake shared library."""
fake_lib_filename = Path(tmp_path) / "fakelib"
- with pytest.raises(Exception):
+ with pytest.raises(Exception): # noqa: B017
load_library(fake_lib_filename)
diff --git a/tests/test_tracker.py b/tests/test_tracker.py
index eaaab346..0d6a86e1 100644
--- a/tests/test_tracker.py
+++ b/tests/test_tracker.py
@@ -30,8 +30,7 @@ def _load_ground_truth_graph() -> dict:
def _get_tracklet(tracks: dict, idx: int) -> list:
"""Get a tracklet by the first object ID"""
- target = [t for t in tracks.values() if t[0] == idx]
- if target:
+ if target := [t for t in tracks.values() if t[0] == idx]:
return target[0]
else:
raise ValueError("Object ID not found.")
@@ -108,6 +107,4 @@ def test_tracker_candidate_graph(test_real_objects, store_graph):
edges = tracker.candidate_graph_edges()
# graph should contain edges
- assert (
- bool(edges) == store_graph
- ), f"Found {len(edges)} edges in candidate graph."
+ assert bool(edges) == store_graph, f"Found {len(edges)} edges in candidate graph."
diff --git a/tests/test_tracklets.py b/tests/test_tracklets.py
index 2ca8d8d7..5a51bd6b 100644
--- a/tests/test_tracklets.py
+++ b/tests/test_tracklets.py
@@ -42,7 +42,7 @@ def test_object_features(properties: dict):
assert obj.n_features == 0
keys = list(properties.keys())
obj.set_features(keys)
- n_keys = sum([np.asarray(p).size for p in properties.values()])
+ n_keys = sum(np.asarray(p).size for p in properties.values())
assert obj.n_features == n_keys
diff --git a/tests/test_utils.py b/tests/test_utils.py
index 164e6564..431f9dde 100644
--- a/tests/test_utils.py
+++ b/tests/test_utils.py
@@ -33,9 +33,7 @@ def _validate_centroids(centroids, objects, scale=None):
obj_as_array = obj_as_array[:, 1:]
# sort the centroids by axis
- centroids = centroids[
- np.lexsort([centroids[:, dim] for dim in range(ndim)][::-1])
- ]
+ centroids = centroids[np.lexsort([centroids[:, dim] for dim in range(ndim)][::-1])]
# sort the objects
obj_as_array = obj_as_array[
@@ -96,9 +94,7 @@ def test_segmentation_to_objects_scale(scale):
def test_assign_class_ID(ndim, nobj):
"""Test mask class_id assignment."""
img, centroids = create_test_image(ndim=ndim, nobj=nobj, binary=False)
- objects = utils.segmentation_to_objects(
- img[np.newaxis, ...], assign_class_ID=True
- )
+ objects = utils.segmentation_to_objects(img[np.newaxis, ...], assign_class_ID=True)
# check that the values match
for obj in objects:
centroid = (int(obj.z), int(obj.y), int(obj.x))[-ndim:]
@@ -112,9 +108,7 @@ def test_regionprops():
"area",
"axis_major_length",
)
- objects = utils.segmentation_to_objects(
- img[np.newaxis, ...], properties=properties
- )
+ objects = utils.segmentation_to_objects(img[np.newaxis, ...], properties=properties)
# check that the properties keys match
for obj in objects:
@@ -167,16 +161,12 @@ def test_update_segmentation_2d(test_segmentation_and_tracks):
@pytest.mark.parametrize("color_by", ["ID", "root", "generation", "fake"])
-def test_update_segmentation_2d_colorby(
- test_segmentation_and_tracks, color_by
-):
+def test_update_segmentation_2d_colorby(test_segmentation_and_tracks, color_by):
"""Test relabeling a 2D-segmentation with track ID."""
in_segmentation, out_segmentation, tracks = test_segmentation_and_tracks
with pytest.raises(ValueError) if color_by == "fake" else nullcontext():
- _ = utils.update_segmentation(
- in_segmentation, tracks, color_by=color_by
- )
+ _ = utils.update_segmentation(in_segmentation, tracks, color_by=color_by)
def test_update_segmentation_3d(test_segmentation_and_tracks):
@@ -260,14 +250,46 @@ def test_tracks_to_napari_ndim_inference(ndim: int):
assert data.shape[-1] == ndim + 2
+def test_napari_to_tracks(sample_tracks):
+ """Test that a napari Tracks layer can be converted to a list of Tracklets.
+
+ First convert tracks to a napari layer, then convert back and compare.
+ """
+
+ data, properties, graph = utils.tracks_to_napari(sample_tracks)
+ tracks = utils.napari_to_tracks(data, properties, graph)
+
+ properties_to_compare = [
+ "ID",
+ "t",
+ "x",
+ "y",
+ # "z", # z-coordinates are different
+ "parent",
+ "label",
+ "state",
+ "root",
+ "is_root",
+ "is_leaf",
+ "start",
+ "stop",
+ "generation",
+ "dummy",
+ "properties",
+ ]
+
+ sample_tracks_dicts = [
+ sample.to_dict(properties_to_compare) for sample in sample_tracks
+ ]
+ tracks_dicts = [track.to_dict(properties_to_compare) for track in tracks]
+ assert sample_tracks_dicts == tracks_dicts
+
+
def test_objects_from_array(test_objects):
"""Test creation of a list of objects from a numpy array."""
obj_arr = np.stack(
- [
- [getattr(obj, k) for k in DEFAULT_OBJECT_KEYS]
- for obj in test_objects
- ],
+ [[getattr(obj, k) for k in DEFAULT_OBJECT_KEYS] for obj in test_objects],
axis=0,
)