-
Notifications
You must be signed in to change notification settings - Fork 24
1. Getting started
In this section, we will learn
- How to install Python using the Anaconda distribution.
- How to install GrainSizeTools.
- How to use Python through Jupyter Notebook or JupyterLab.
- How to interact with the script.
Note
Although this script does not require any prior knowledge of Python programming language, a general introduction to Python can be found at the following web link: https://marcoalopez.github.io/Python_course/
GrainSizeTools requires Python 3, the Python scientific libraries NumPy SciPy, Pandas, Matplotlib, and JupyterLab. If you have no previous experience with Python, we recommend downloading and installing the Anaconda Python distribution which is free and contains all the necessary scientific packages (> 5 GB disc space). There are versions for Windows, MacOS and Linux.
https://docs.anaconda.com/free/anaconda/install/
Tip
If you have limited disk space, there is a distribution called Miniconda which installs only the Python packages you need. If you prefer to go to this route, click here for detailed instructions.
Once Anaconda is installed, launch the Anaconda Navigator and you will see that you have installed various scientifically oriented Integrated Development Systems (IDEs), including JupyterLab and the classic Jupyter Notebook. Clicking on any of these will open the corresponding application in your default web browser. The GrainSizeTools documentation is written assuming that you will be working using Jupyter Notebooks.
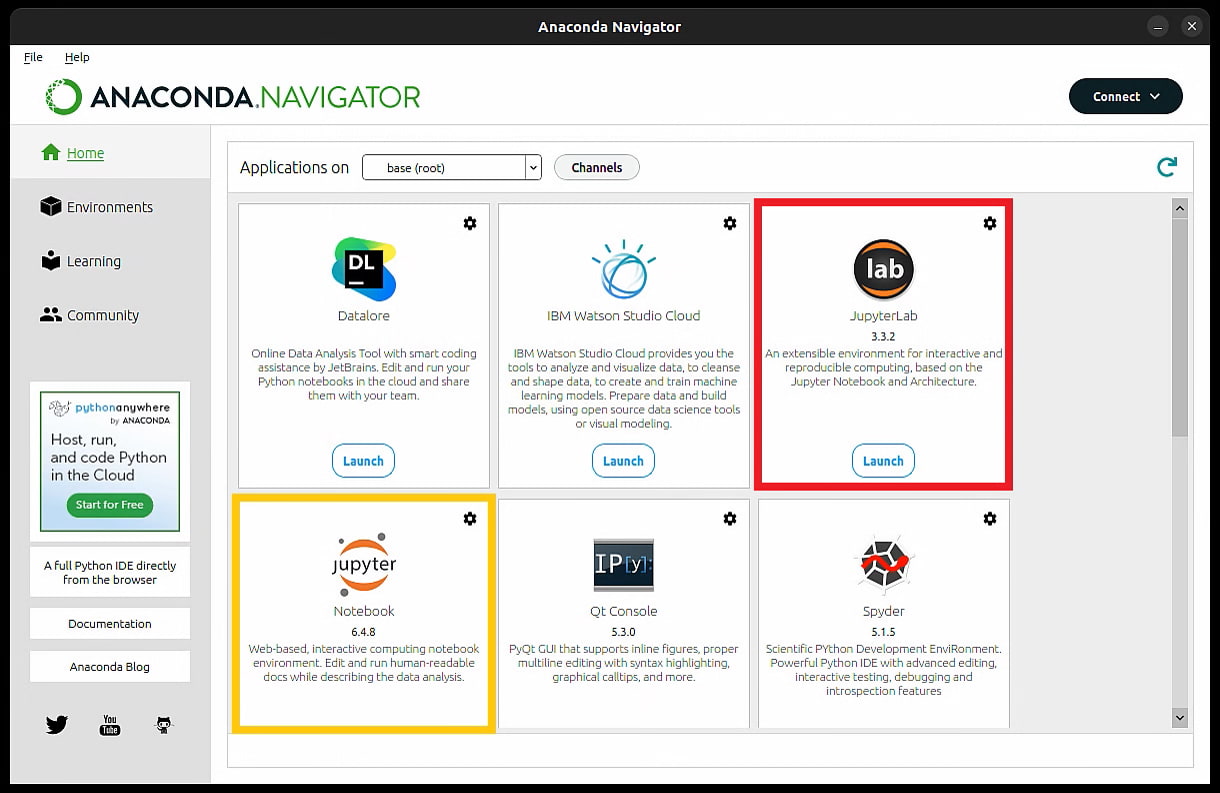
The appearance of the Anaconda navigator. Framed in red and orange are JupyterLab (preferred option) and Jupyter Notebooks (both should be installed by default in Anaconda). JupyterLab is the next generation of the Classic Jupyter Notebook application interface.
Tip
Using a dedicated application to work with Jupyter Notebooks
If you prefer to use a dedicated application instead of opening Jupyter Notebooks in your browser, there are several alternatives. Here we will mention two free alternatives:
- JupyterLab desktop: https://github.com/jupyterlab/jupyterlab-desktop/releases
This is a cross-platform desktop application for JupyterLab. It is the same application that opens in the browser but in an encapsulated application. You can find the user guide at the following link https://github.com/jupyterlab/jupyterlab-desktop/blob/master/user-guide.md. If you are a beginner, this is the easy road.
- Visual Studio Code (a.k.a. Vscode): https://code.visualstudio.com/
This is a free code editor that can be used with various programming languages including Python and supports Jupyter Notebooks via extensions. As an advantage over vanilla JupyterLab, it has a handy variable browser. More detailed instructions on how to use Jupyter Notebooks in Vscode at the following link https://code.visualstudio.com/docs/datascience/jupyter-notebooks
Note that both applications require Python to be installed on your operating system, i.e. it does not exempt you from the step of installing Python using Anaconda or any other distribution.
Once Python is installed, the next step is to download GrainSizeTools. Click on the download link below (there is also a direct link on the GrainSizeTools website).
https://github.com/marcoalopez/GrainSizeTools/releases
and download the latest version of the script by clicking on the zip file named grain_size_tools_v2024.03.20.zip (numbers may change!) as shown below
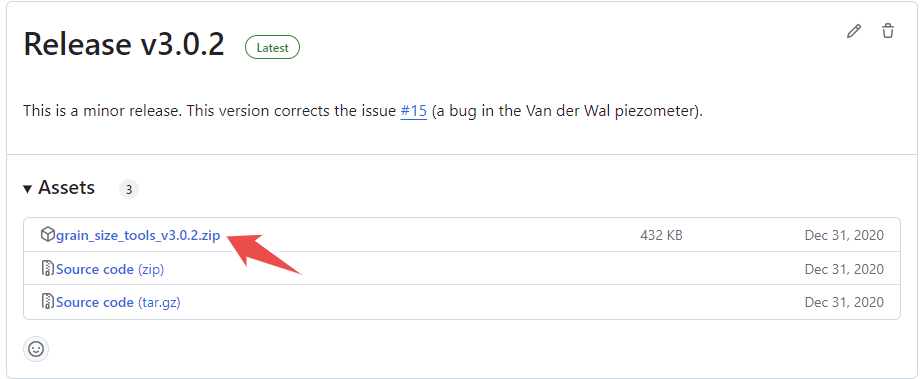
Unzip the file and save the GrainSizeTools folder to a location of your choice. The GrainSizeTools folder contains various Python files (.py), a folder named DATA with a CSV file inside, and various Jupyter Notebooks (.ipynb files) that are templates for doing different types of grain size data analysis.
To improve the reproducibility of the grain size studies, we suggest working with the Jupyter Notebooks templates provided within the script, especially if you have no previous experience with the Python programming language. A Jupyter Notebook is a document that can contain executable code, equations, visualisations and narrative text so that the user bring together data, code, figures and prose in a single document. This is ideal for generating reports and including them as supplementary material in your publications so that any researcher can reproduce your results.
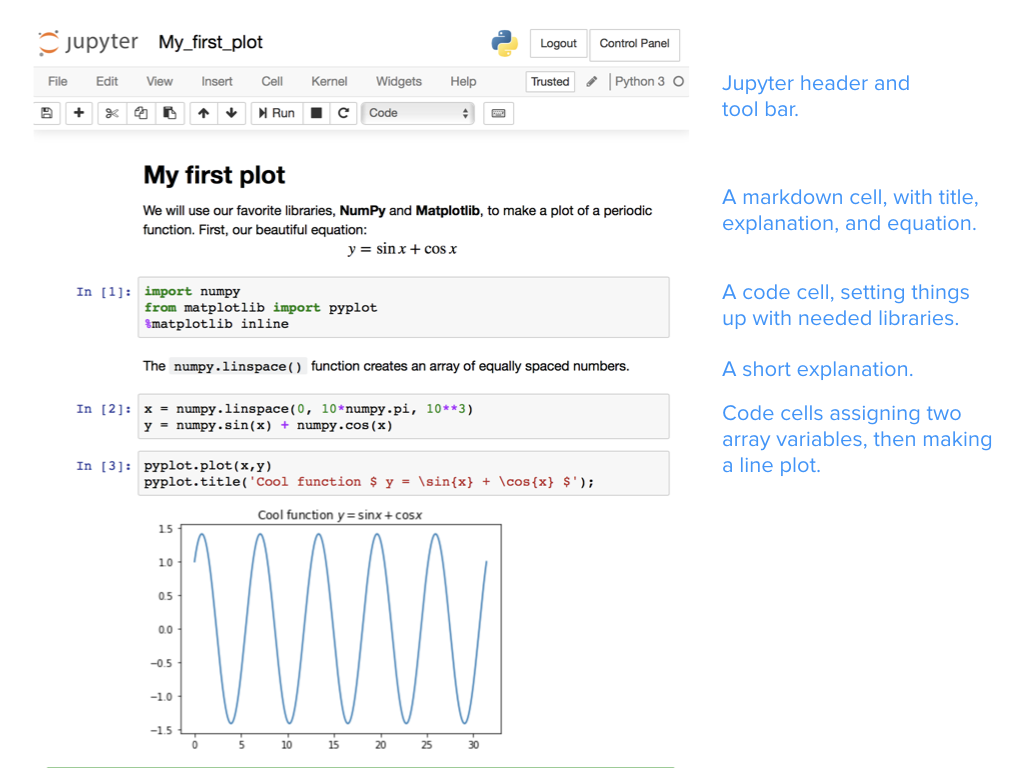
A Jupyter notebook, starting with a markdown cell containing a title and an explanation (including an equation rendered with LaTeX). Three code cells produce the final inline plot. Source: Teaching and Learning with Jupyter https://jupyter4edu.github.io/jupyter-edu-book/
Note
A detailed explanation of how Jupyter Notebooks work is beyond the scope of this wiki page. The aim here is to teach the minimum necessary for a user to be able to use the Jupyter Notebook templates that come with the script. In any case, it is recommended that the user becomes familiar with the use of Jupyter Notebooks. There are excellent tutorials on the web explaining how to use Jupyter Notebooks, here are some suggestions::
-
https://www.youtube.com/watch?v=HW29067qVWk This is a video by Corey Schafer that explains in a clear, entertaining and concise way how to install and use a Jupyter Notebook, the usage part starts at about 4:20. For the tutorial, Corey uses the classic Jupyter Notebook application instead of JupyterLab, but it works similarly.
-
TODO
When you first open JupyterLab, you should see something similar to this
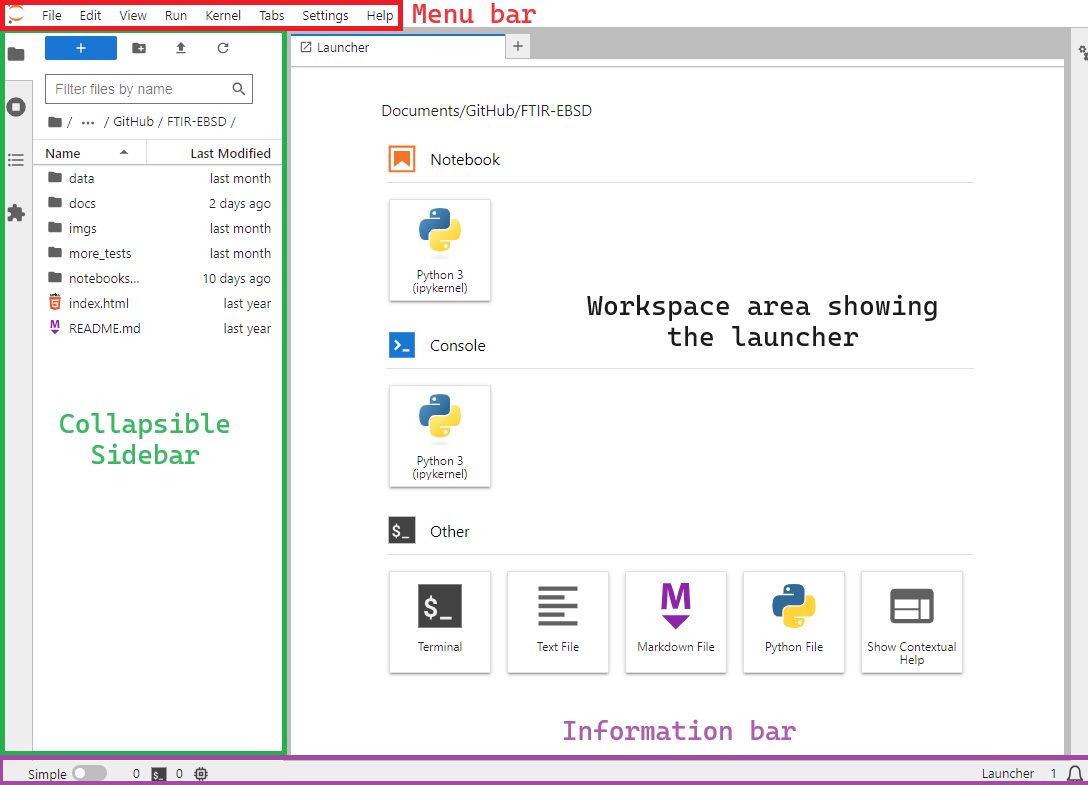
you should familiarise yourself with the following elements:
-
The Menu Bar at the top of the screen, highlighted in red, with familiar drop-down menus suc as “File”, “Edit”, “View”, etc.
-
The Workspace Area currently displaying the Launcher. This allows users to access the Console, the Terminal or create new Notebooks, Python, Text, or Markdown files.
-
The Collapsible Sidebar, highlighted in green on the left, contains a File Browser, along with other items such as running tabs and kernels information, notebook table of contents, and the Extensions Manager.
-
The Information Bar highlighted in purple and located below.
At this point, use the launcher to create your first Jupyter notebook.
When you create a notebook, you'll find an empty cell at the top. These cells can be used for writing code, text (which is called markdown), or raw content. When you click in a cell, it turns white with a blinking cursor to indicate that it's ready for editing. The type of cell is shown in the top bar of the current tab. You can change the cell type at any time.
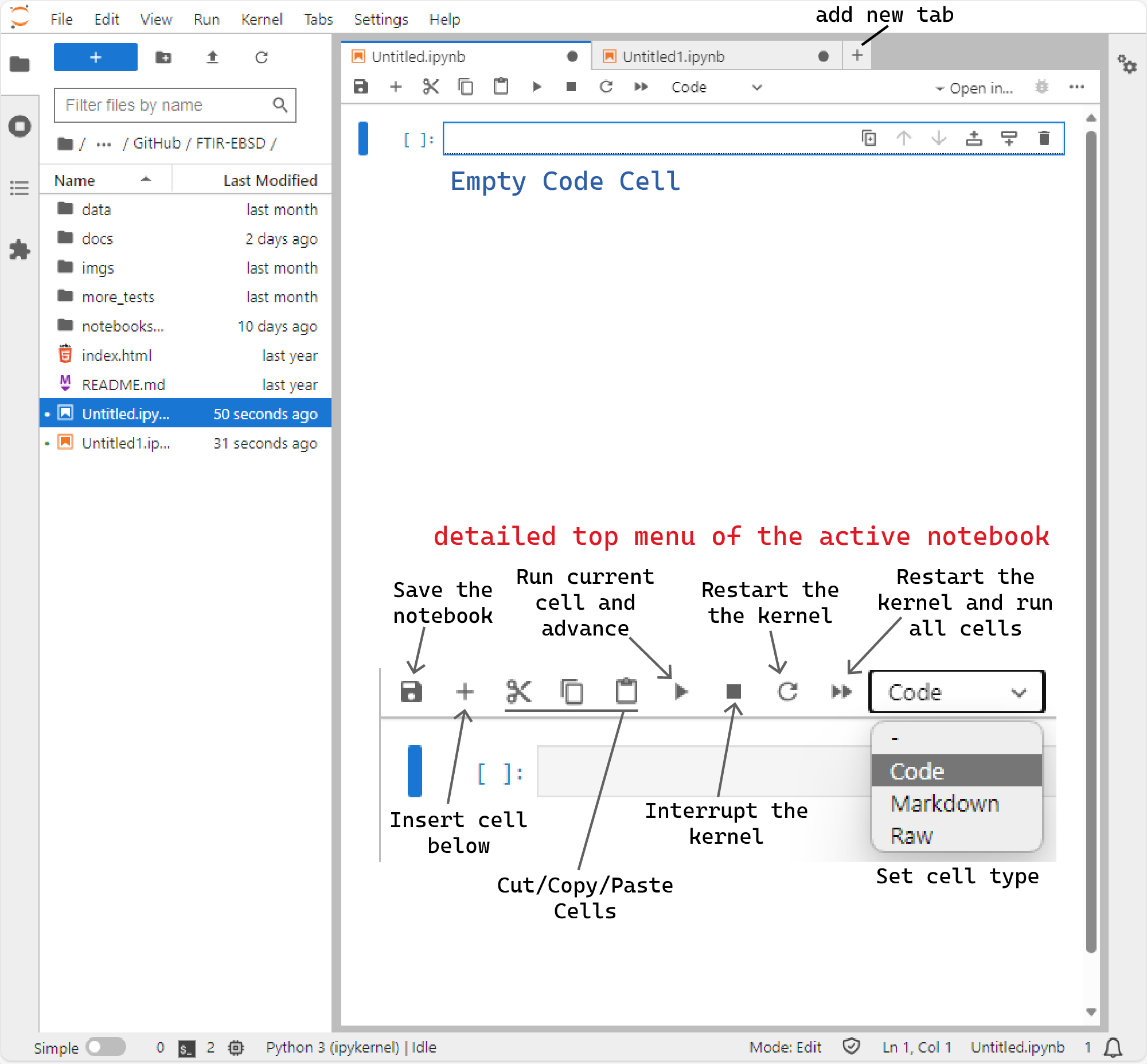
JupyterLab interface at a glance. More info at https://jupyterlab.readthedocs.io/en/latest/user/interface.html#the-jupyterlab-interface
If it's a code cell, you can write Python code in it. When you run the cell by pressing Shift+Enter the code gets executed, and any results appear just below the cell. Alternatively, to can run a cell clicking on the Play icon in the top bar of the notebook. The Shift+Enter action will create a new empty cell just below it. If it is a markdown cell, you can use Markdown to write your own annotations. When you're ready, press Shift+Enter and the text will be rendered and appear fully formatted. It will also create a new empty cell below it. Repeat this as many times as you need.
When you open an edited notebook, for example the notebook templates that come with the script, you can edit any cell by double-clicking on the cell you want to modify and executing it once you have finished editing it.
To open...TODO...using the collapsible sidebar or File > Open from Path, the cells will already be created, and
The script consists of seven Python files, listed below, which must be in the same folder.
-
GrainSizeTools.py: This file imports all the modules needed for the script to work. -
averages.py: This module contains a set of functions for calculating different types of averages and margins of error. -
plot.py: This module contains a set of functions for generating different types of ad hoc plots used by the script. -
stereology.py: This module contains a set of functions to approximate true grain size distributions from sectional measurements using stereological methods. -
piezometric_database.py: This file contains the database of piezometers the script uses. -
template.py: This file contains the default (Matplotlib) parameters used by the script to generate plots. -
get.py: This file contains the welcome message of the script.
GrainSizeTools.py is the only executed Python file in the Jupyter Notebook templates, as it takes care of importing all the other modules of the script.
TODO -> image
TODO...To access the methods of each module, write in a code cell the name of the module, followed by a dot and then the name of the method (module.method()), for example:
averages.amean()We will see the functionality of all the methods in more detail in the examples shown in this wiki. TODO
In the GrainSizeTools folder, you'll find a subfolder called DATA. This is where you should store any grain size data you want to analyse. While it's not strictly necessary, it is a desirable for reproducibility. In addition, the templates assume by default that all the data to be processed is located in this folder, although this can easily be changed.
Important
The GrainSizeTools script is not intended to deal with microscopic images but to quantify and visualize grain size populations and estimate stresses via paleopiezometers. It is, therefore, necessary to measure the grain diameters or the cross-sectional of the grains in advance and store them in a txt/CSV/excel file.
If the source of your data are images, we strongly recommend using the ImageJ application or one of its different flavours (see here). ImageJ-type applications are public domain image processing programs widely used in scientific research and run on all operating systems. This wiki includes a short tutorial on how to measure the areas of the grain profiles with ImageJ. The combined use of ImageJ and GrainSizeTools script is intended to ensure that all data processing steps are done through free and open-source programs/scripts that run under any operating system.
If you are dealing with EBSD data instead, we encourage you to use the MTEX toolbox for grain reconstruction and extracting the grain size data.
The GrainSizeTools folder contains three different notebook templates depending on the type of study you want to perform:
- quantification of grain size distributions (
grainsize_pop_template.ipynb) - approximation of grain size distributions using stereological methods (
stereology_template.ipynb), - paleopiezometry (
paleopiezometry_template.ipynb).
The notebooks contain all the necessary instructions on how to use them through practical examples. The user only has to change some parameters in the cells to adapt them to their study case and delete everything that is not needed. Once this is done, all the data and outputs will be contained in a single folder and the data handling procedures in a single fully reproducible document that can be exported to other formats (PDF, HTML, etc.) if necessary. When using this workflow, you should ideally copy the entire GrainSizeTool folder (<<1 MB), i.e. the code, the subfolder structure and the notebooks, for each grain size study you perform.
Once you have everything installed, open JupyterLab or Jupyter Notebooks and in the file manager go to the location where the script and templates are. Open the Jupyter template you want to use and follow the instructions in the template. That's it.
Caution
In your notebook, you can run cells in any order by selecting them and pressing Shift+Enter. But, when you share your notebook, it's best for reproducibility to run cells in the order they're shown. To do this, you can restart the kernel and then go to the menu bar and click on Run > Run All (the top menu of the notebook also has two icons for this purpose). This helps prevent unexpected issues if you're using variables between cells and improve reproducibility. Just be cautious when changing the order.
TODO