R/Medicine was virtual this year, so I finally got to attend. Both R/Med and csv,conf used Crowdcast - my favourite of the many virtual conference platforms I've experienced this year. R/Med was really well organized, with great speakers and a fantastic mix of topics from R, statistics, and clinical studies.
There was a lot of information packed into this 2.5-day conference, so I haven't had time to process everything, but here's a quick list of interesting things that I need to read more about later.
Currently the recordings are available on Crowdcast for registrants, but I've tried to link to open sources below. Check out @r_medicine or #RMedicine2020 for recaps and conversations.
-
Tidymodels/machine learning workshop - this was fantastic, now just need to use this again so I can remember it! We went through an example of building a supervised classification model. Website has links to all the slides and exercises.
-
Reading material:
- Double dipping in statistical analysis - what is double dipping, and how to fix by conditioning on hypothesis selection procedure. Preprint is still in preparation, but Twitter thread gives a good summary.
- HealthyR - training & resources for health data analysis
- Arcus Education at Children's Hospital of Philadelphia - more training & resources
-
Shiny apps:
- {shinyviz} - template Shiny app for summarizing & visualizing categorical data; customize for your own data
- {shinyfit} - template Shiny app for linear/logistic/etc. regression analysis; customize for your own data
- tidyCDISC - Shiny app to explore & visualize ADaM-format clinical trial data; slides here; will be on GitHub "soon"
- {nDSPA} - Shiny app & package for working with spatial omics data
- READi Tool - Shiny app for evaluating real-world data - creates custom PubMed search from input; summary here; still in beta testing (app seems to be down as of 9/4/2020)
-
Packages - general R/R Markdown/Shiny:
- {listdown} - programmatically generate R Markdown files from named lists
- {dbplyr} - do data manipulation on SQL server, then just pull what you need into R for analysis
- {drake} - plan your workflow so you don't have to keep repeating the computationally intensive parts
- {pkgdown} - build a website for your R package (i.e. pkg documentation as knowledge repository)
- Docker & {holepunch} - share research compendia on GitHub
- {shinyviz} - template Shiny app for summarizing & visualizing categorical data
- {shinyfit} - template Shiny app for linear/logistic/etc. regression analysis
- {golem} - opinionated framework for building robust Shiny apps
- {pool} to make a separate data layer for your workflow/Shiny app
-
Packages - Publications/communication
- {gtsummary} - make publication-ready analytical and summary tables. Example:

- {finalfit} - make nice tables for publication; similar to {gtsummary}
-
Packages - Clinical data
- {survminer} - make Kaplan-Meier curves with ggplot
- {ggconsort} - programatically generate CONSORT diagrams; still in development
- {foreceps} - processing & wrangle ADaM-format clinical trial data; will be available in a few weeks
- {collaboratoR} - work with REDCap data in R
- {redcapR} - interact with the REDCap API
-
Packages - Other
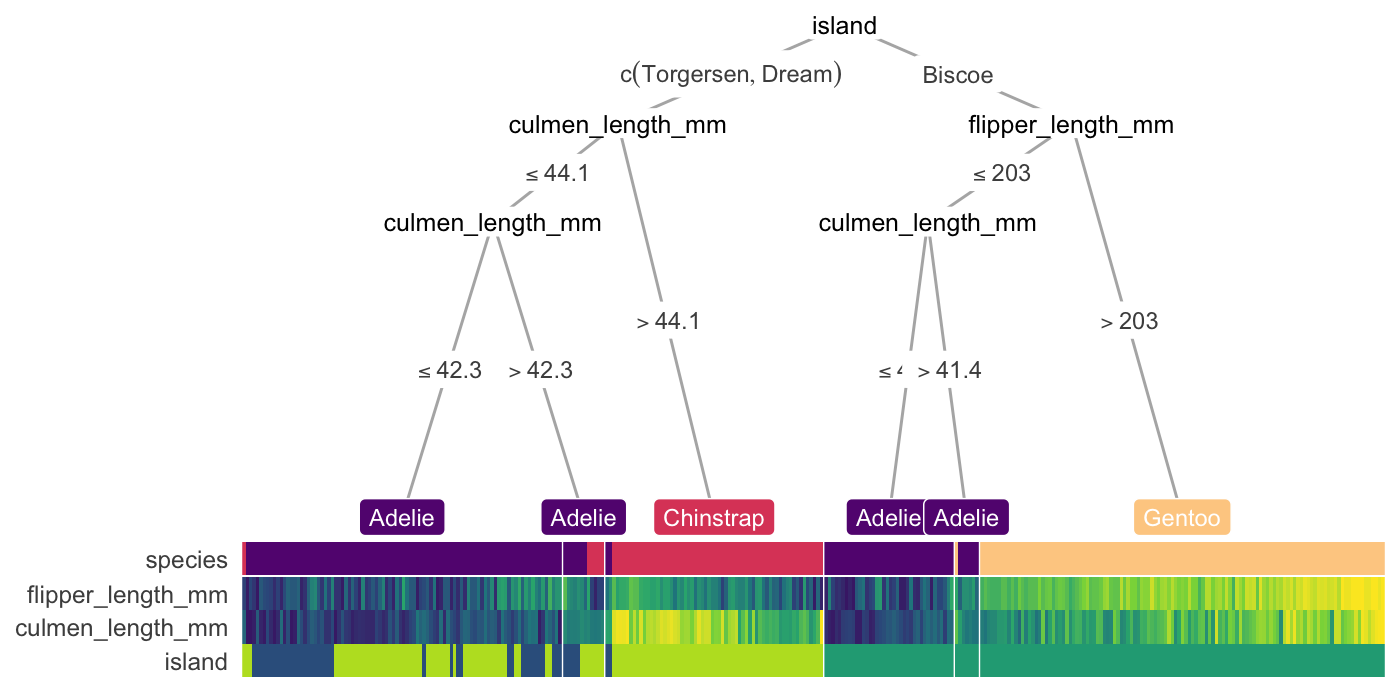
- {treeheatr} - decision tree + heatmap visualization for better interpretability. Example:
- {runcharter} - plot & analyze run charts to track changes over time. Example:

- {twilio} - connect to Twilio API to send text messages from R
- {treeheatr} - decision tree + heatmap visualization for better interpretability. Example: