diff --git a/DESCRIPTION b/DESCRIPTION
index 064ec65..2dd6cc8 100644
--- a/DESCRIPTION
+++ b/DESCRIPTION
@@ -1,6 +1,6 @@
Package: easyalluvial
Title: Generate Alluvial Plots with a Single Line of Code
-Version: 0.3.1.900
+Version: 0.3.2
Authors@R: person( "Bjoern", "Koneswarakantha", role = c("aut","cre"), email = "datistics@gmail.com", comment = c(ORCID = "0000-0003-4585-7799") )
URL: https://github.com/erblast/easyalluvial/
Description: Alluvial plots are similar to sankey diagrams and visualise categorical data
diff --git a/NEWS.md b/NEWS.md
index 74c2b8a..b801fe7 100644
--- a/NEWS.md
+++ b/NEWS.md
@@ -1,3 +1,7 @@
+# Version 0.3.2 submitted to CRAN 20231207
+- check for vip pkg installation before executing example of alluvial_model_response_parsnip
+- option for using geom_text instead of geom_label (by https://github.com/gernophil)
+
# Version 0.3.1 submitted to CRAN 20220704
- cleanup package dependencies
diff --git a/R/alluvial_model_response.R b/R/alluvial_model_response.R
index d9f24be..d75396b 100644
--- a/R/alluvial_model_response.R
+++ b/R/alluvial_model_response.R
@@ -236,8 +236,8 @@ tidy_imp = function(imp, df, .f = max, resp_var = NULL){
#' \code{\link[easyalluvial]{manip_bin_numerics}}, while leaving categoric
#' variables unchanged. The number of bins for each numeric variable is set to
#' bins -2. Next the median is picked for each of the bins and the min and the
-#' max value is added for each numeric variable So that we get { median(bin) X
-#' bins -2, max, min} for each numeric variable. Then all possible combinations
+#' max value is added for each numeric variable So that we get (median(bin) X
+#' bins -2, max, min) for each numeric variable. Then all possible combinations
#' between those values and the categoric factor levels are created. The total
#' number of all possible combinations defines the range of the data space. The
#' values of the remaining variables are set to mode(factors) or
diff --git a/R/alluvial_wide.R b/R/alluvial_wide.R
index d2b0c08..6c60131 100644
--- a/R/alluvial_wide.R
+++ b/R/alluvial_wide.R
@@ -395,7 +395,7 @@ alluvial_wide = function( data
data_key = data_new %>%
mutate( alluvial_id = manip_factor_2_numeric(alluvial_id) ) %>%
- left_join( data_alluvial ) %>%
+ left_join( data_alluvial, relationship = "many-to-many" ) %>%
select( - fill_flow, -fill_value, -fill ) %>%
spread( key = x, value = value ) %>%
select( one_of(id_str, variables, 'alluvial_id', 'n' ) ) %>%
diff --git a/README.Rmd b/README.Rmd
index 886bfe9..3b44fd4 100644
--- a/README.Rmd
+++ b/README.Rmd
@@ -17,7 +17,6 @@ knitr::opts_chunk$set(
# easyalluvial
-[](https://ci.appveyor.com/project/erblast/easyalluvial)
[](https://github.com/erblast/easyalluvial/actions)
[](https://codecov.io/github/erblast/easyalluvial?branch=master)
[](https://CRAN.R-project.org/package=easyalluvial)
diff --git a/README.md b/README.md
index 7630333..c360919 100644
--- a/README.md
+++ b/README.md
@@ -3,8 +3,6 @@
# easyalluvial
-[](https://ci.appveyor.com/project/erblast/easyalluvial)
[](https://github.com/erblast/easyalluvial/actions)
[
-- automatically transforms numerical to categorical data
-- helper functions for variable selection
-- convenient parameters for coloring and ordering
-- marginal histograms
-- **model agnostic partial dependence and model response alluvial
- plots with 4 dimensions**
-- **[interactive plots with `easyalluvial` and
- `parcats`](https://erblast.github.io/parcats/articles/parcats.html)**
+- plot alluvial graph with a single line of code of a given dataframe
+- support for wide and long data format [(wiki, wide vs. long/narrow
+ data)](https://en.wikipedia.org/wiki/Wide_and_narrow_data)
+- automatically transforms numerical to categorical data
+- helper functions for variable selection
+- convenient parameters for coloring and ordering
+- marginal histograms
+- **model agnostic partial dependence and model response alluvial plots
+ with 4 dimensions**
+- **[interactive plots with `easyalluvial` and
+ `parcats`](https://erblast.github.io/parcats/articles/parcats.html)**
## Installation
@@ -60,19 +58,20 @@ install.packages('easyalluvial')
### Development Version
``` r
+
# install.packages("devtools")
devtools::install_github("erblast/easyalluvial")
```
## Documentation
-- [pkgdown website](https://erblast.github.io/easyalluvial/)
- - [Data Exploration with Alluvial
- Plots](https://erblast.github.io/easyalluvial/articles/data_exploration.html)
- - [Visualising Model
- Response](https://erblast.github.io/easyalluvial/articles/model_response.html)
- - [Interactive Plots with
- parcats](https://erblast.github.io/easyalluvial/articles/parcats.html)
+- [pkgdown website](https://erblast.github.io/easyalluvial/)
+ - [Data Exploration with Alluvial
+ Plots](https://erblast.github.io/easyalluvial/articles/data_exploration.html)
+ - [Visualising Model
+ Response](https://erblast.github.io/easyalluvial/articles/model_response.html)
+ - [Interactive Plots with
+ parcats](https://erblast.github.io/easyalluvial/articles/parcats.html)
## Examples
@@ -86,6 +85,7 @@ suppressPackageStartupMessages( require(easyalluvial) )
#### Sample Data
``` r
+
knitr::kable( head(mtcars2) )
```
@@ -102,13 +102,14 @@ knitr::kable( head(mtcars2) )
Continuous Variables will be automatically binned as follows.
-- High, High (HH)
-- Medium, High (MH)
-- Medium (M)
-- Medium, Low (ML)
-- Low, Low (LL)
+- High, High (HH)
+- Medium, High (MH)
+- Medium (M)
+- Medium, Low (ML)
+- Low, Low (LL)
``` r
+
alluvial_wide( data = mtcars2
, max_variables = 5
, fill_by = 'first_variable' )
@@ -124,18 +125,19 @@ alluvial_wide( data = mtcars2
knitr::kable( head(quarterly_flights) )
```
-| tailnum | carrier | origin | dest | qu | mean\_arr\_delay |
-|:------------------|:--------|:-------|:-----|:----|:-----------------|
-| N0EGMQ LGA BNA MQ | MQ | LGA | BNA | Q1 | on\_time |
-| N0EGMQ LGA BNA MQ | MQ | LGA | BNA | Q2 | on\_time |
-| N0EGMQ LGA BNA MQ | MQ | LGA | BNA | Q3 | on\_time |
-| N0EGMQ LGA BNA MQ | MQ | LGA | BNA | Q4 | on\_time |
-| N11150 EWR MCI EV | EV | EWR | MCI | Q1 | late |
-| N11150 EWR MCI EV | EV | EWR | MCI | Q2 | late |
+| tailnum | carrier | origin | dest | qu | mean_arr_delay |
+|:------------------|:--------|:-------|:-----|:----|:---------------|
+| N0EGMQ LGA BNA MQ | MQ | LGA | BNA | Q1 | on_time |
+| N0EGMQ LGA BNA MQ | MQ | LGA | BNA | Q2 | on_time |
+| N0EGMQ LGA BNA MQ | MQ | LGA | BNA | Q3 | on_time |
+| N0EGMQ LGA BNA MQ | MQ | LGA | BNA | Q4 | on_time |
+| N11150 EWR MCI EV | EV | EWR | MCI | Q1 | late |
+| N11150 EWR MCI EV | EV | EWR | MCI | Q2 | late |
#### Plot
``` r
+
alluvial_long( quarterly_flights
, key = qu
, value = mean_arr_delay
@@ -159,6 +161,7 @@ alluvial_wide( data = mtcars2
### Interactive Graphs
``` r
+
suppressPackageStartupMessages( require(parcats) )
p = alluvial_wide(mtcars2, max_variables = 5)
@@ -166,10 +169,15 @@ p = alluvial_wide(mtcars2, max_variables = 5)
parcats(p, marginal_histograms = TRUE, data_input = mtcars2)
```
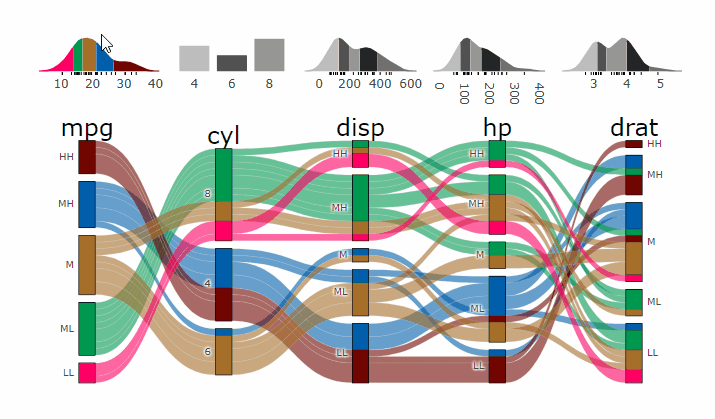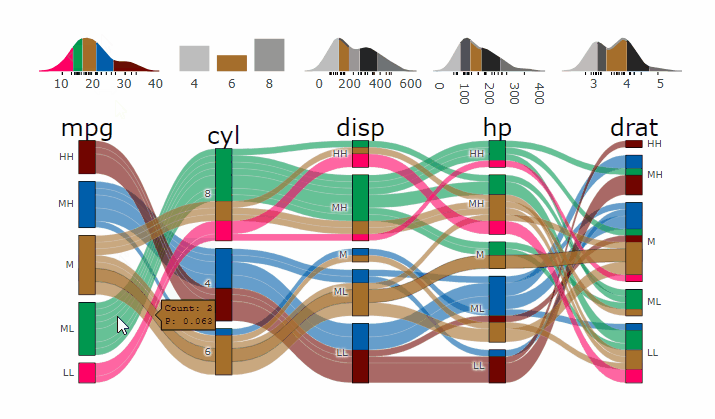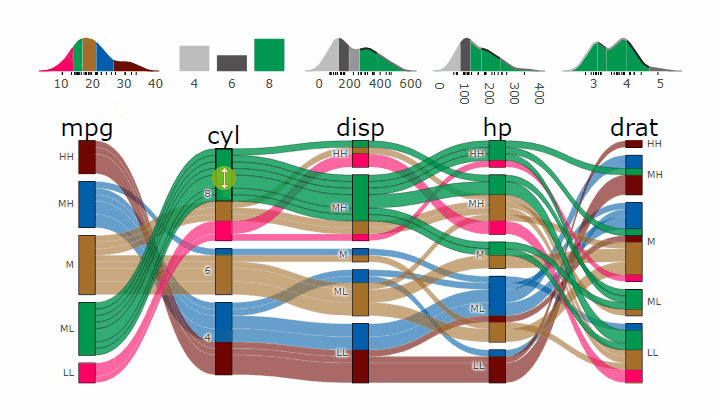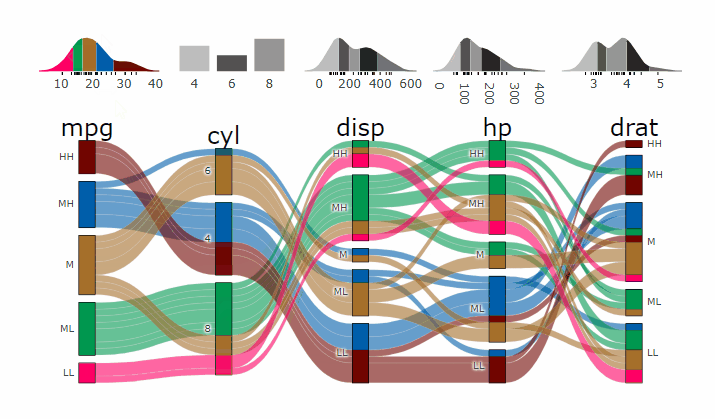
-
+
+
+demo
+
-- **[Live
- Widget](https://erblast.github.io/parcats/articles/parcats.html)**
+- **[Live
+ Widget](https://erblast.github.io/parcats/articles/parcats.html)**
### Partial Dependence Alluvial Plots
@@ -190,14 +198,15 @@ in the end. Each of the possible combinations is plotted as a flow which
is coloured by the bin corresponding to the average model response
generated by that particular combination.
-- [more on partial dependence plots
- (ebook)](https://christophm.github.io/interpretable-ml-book/)
-- [Tutorial](https://www.datisticsblog.com/2019/04/visualising-model-response-with-easyalluvial/)
+- [more on partial dependence plots
+ (ebook)](https://christophm.github.io/interpretable-ml-book/)
+- [Tutorial](https://www.datisticsblog.com/2019/04/visualising-model-response-with-easyalluvial/)
`easyalluvial` contains wrappers for `parsnip` and `caret` models.
Custom Wrappers for other models can easily be created.
``` r
+
df = select(mtcars2, -ids)
m = parsnip::rand_forest(mode = "regression") %>%
@@ -216,11 +225,12 @@ p_grid = add_marginal_histograms(p, df, plot = F) %>%
### Interactive Partial Dependence Plot
``` r
+
parcats(p, marginal_histograms = TRUE, imp = TRUE, data_input = df)
```
-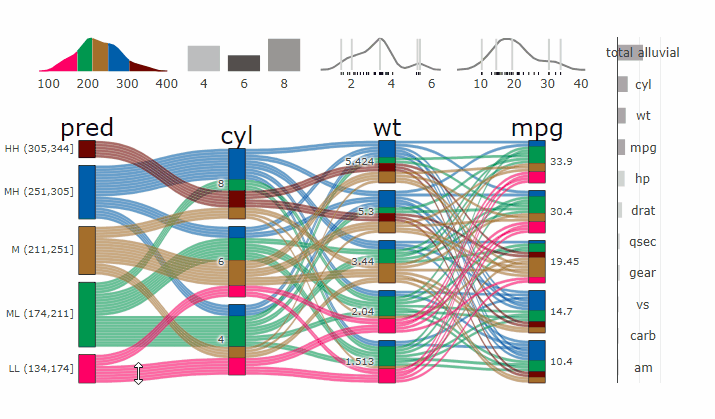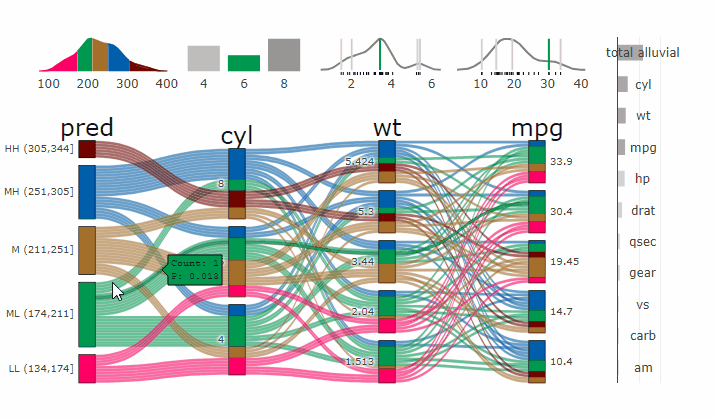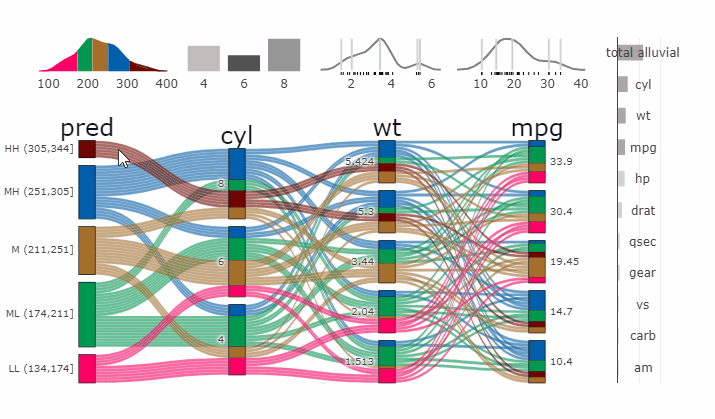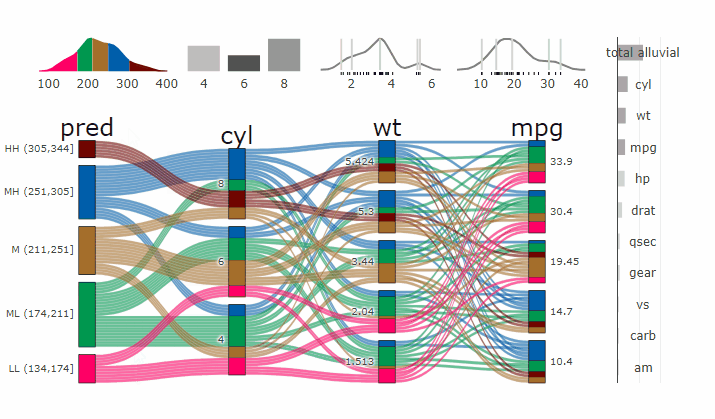
-- **[Live
+ -
+**[Live
Widget](https://erblast.github.io/parcats/articles/parcats.html)**
# ClinicoPath {jamovi} Module
@@ -232,7 +242,7 @@ interface for doing statistics with `R`.
# Similar Packages
-- [`ggalluvial`](https://github.com/corybrunson/ggalluvial/)
-- [`alluvial`](https://github.com/mbojan/alluvial)
-- [`networkD3`](https://github.com/christophergandrud/networkD3)
-- [`ggbump`](https://github.com/davidsjoberg/ggbump)
+- [`ggalluvial`](https://github.com/corybrunson/ggalluvial/)
+- [`alluvial`](https://github.com/mbojan/alluvial)
+- [`networkD3`](https://github.com/christophergandrud/networkD3)
+- [`ggbump`](https://github.com/davidsjoberg/ggbump)
diff --git a/appveyor.yml b/appveyor.yml
deleted file mode 100644
index c6c1438..0000000
--- a/appveyor.yml
+++ /dev/null
@@ -1,45 +0,0 @@
-# DO NOT CHANGE the "init" and "install" sections below
-
-# Download script file from GitHub
-init:
- ps: |
- $ErrorActionPreference = "Stop"
- Invoke-WebRequest http://raw.github.com/krlmlr/r-appveyor/master/scripts/appveyor-tool.ps1 -OutFile "..\appveyor-tool.ps1"
- Import-Module '..\appveyor-tool.ps1'
-
-install:
- ps: Bootstrap
-
-cache:
- - C:\RLibrary
-
-# Adapt as necessary starting from here
-
-build_script:
- - travis-tool.sh install_deps
-
-test_script:
- - travis-tool.sh run_tests
-
-on_failure:
- - 7z a failure.zip *.Rcheck\*
- - appveyor PushArtifact failure.zip
-
-artifacts:
- - path: '*.Rcheck\**\*.log'
- name: Logs
-
- - path: '*.Rcheck\**\*.out'
- name: Logs
-
- - path: '*.Rcheck\**\*.fail'
- name: Logs
-
- - path: '*.Rcheck\**\*.Rout'
- name: Logs
-
- - path: '\*_*.tar.gz'
- name: Bits
-
- - path: '\*_*.zip'
- name: Bits
diff --git a/cran-comments.md b/cran-comments.md
index 17d0f5b..c13916a 100644
--- a/cran-comments.md
+++ b/cran-comments.md
@@ -1,12 +1,19 @@
# Resubmission after being archived on CRAN
-clean up package dependencies
+All suggested dependencies were already controlled for, meaning there was a
+check in place that would raise an error with a message to install the missing
+package. Those are limited to functions most users will not use.
+
+All unit tests would skip if suggested packages were missing.
+
+One example missed a check for a suggested package installation which was added:
+
+- check for vip pkg installation before executing example of alluvial_model_response_parsnip
## Test Environments
-* local macOS M1 R 4.2.1
-* Appveyor x86_64-w64-mingw32/x64 (64-bit) R 4.2.1
-* github actions macos-latest R 4.2.1
-* WinBuilder R 4.2.1
+* local macOS M1 R 4.3.2
+* github actions macos-latest R 4.3.2
+* WinBuilder R 4.3.2
* WinBuilder R devel
coverage -> 96 %
diff --git a/man/figures/README-plot_long-1.png b/man/figures/README-plot_long-1.png
index 5983a51..c343c89 100644
Binary files a/man/figures/README-plot_long-1.png and b/man/figures/README-plot_long-1.png differ
diff --git a/man/figures/README-unnamed-chunk-3-1.png b/man/figures/README-unnamed-chunk-3-1.png
index 041c35f..c81f208 100644
Binary files a/man/figures/README-unnamed-chunk-3-1.png and b/man/figures/README-unnamed-chunk-3-1.png differ
diff --git a/man/figures/README-unnamed-chunk-5-1.png b/man/figures/README-unnamed-chunk-5-1.png
index e9bea3f..6cc3ba0 100644
Binary files a/man/figures/README-unnamed-chunk-5-1.png and b/man/figures/README-unnamed-chunk-5-1.png differ
diff --git a/man/figures/README-wide_plot-1.png b/man/figures/README-wide_plot-1.png
index 2b60bf4..c69e2a3 100644
Binary files a/man/figures/README-wide_plot-1.png and b/man/figures/README-wide_plot-1.png differ
diff --git a/man/get_data_space.Rd b/man/get_data_space.Rd
index 6d132ce..54108b5 100644
--- a/man/get_data_space.Rd
+++ b/man/get_data_space.Rd
@@ -41,8 +41,8 @@ It selects a the top most important variables based on the degree
\code{\link[easyalluvial]{manip_bin_numerics}}, while leaving categoric
variables unchanged. The number of bins for each numeric variable is set to
bins -2. Next the median is picked for each of the bins and the min and the
- max value is added for each numeric variable So that we get { median(bin) X
- bins -2, max, min} for each numeric variable. Then all possible combinations
+ max value is added for each numeric variable So that we get (median(bin) X
+ bins -2, max, min) for each numeric variable. Then all possible combinations
between those values and the categoric factor levels are created. The total
number of all possible combinations defines the range of the data space. The
values of the remaining variables are set to mode(factors) or
diff --git a/tests/testthat/_snaps/plot_imp/plot-imp.new.svg b/tests/testthat/_snaps/plot_imp/plot-imp.new.svg
deleted file mode 100644
index b82ec09..0000000
--- a/tests/testthat/_snaps/plot_imp/plot-imp.new.svg
+++ /dev/null
@@ -1,106 +0,0 @@
-
-
diff --git a/tests/testthat/_snaps/plot_imp/plot-imp.svg b/tests/testthat/_snaps/plot_imp/plot-imp.svg
deleted file mode 100644
index bf7144f..0000000
--- a/tests/testthat/_snaps/plot_imp/plot-imp.svg
+++ /dev/null
@@ -1,106 +0,0 @@
-
-
diff --git a/tests/testthat/_snaps/plot_marginal_histograms/mod-cat-pred-train.new.svg b/tests/testthat/_snaps/plot_marginal_histograms/mod-cat-pred-train.new.svg
deleted file mode 100644
index 454ecb2..0000000
--- a/tests/testthat/_snaps/plot_marginal_histograms/mod-cat-pred-train.new.svg
+++ /dev/null
@@ -1,102 +0,0 @@
-
-
diff --git a/tests/testthat/_snaps/plot_marginal_histograms/mod-cat-pred-train.svg b/tests/testthat/_snaps/plot_marginal_histograms/mod-cat-pred-train.svg
deleted file mode 100644
index 4da40f5..0000000
--- a/tests/testthat/_snaps/plot_marginal_histograms/mod-cat-pred-train.svg
+++ /dev/null
@@ -1,102 +0,0 @@
-
-
diff --git a/tests/testthat/_snaps/plot_marginal_histograms/mod-cat-pred.new.svg b/tests/testthat/_snaps/plot_marginal_histograms/mod-cat-pred.new.svg
deleted file mode 100644
index 434399f..0000000
--- a/tests/testthat/_snaps/plot_marginal_histograms/mod-cat-pred.new.svg
+++ /dev/null
@@ -1,76 +0,0 @@
-
-
diff --git a/tests/testthat/_snaps/plot_marginal_histograms/mod-cat-pred.svg b/tests/testthat/_snaps/plot_marginal_histograms/mod-cat-pred.svg
deleted file mode 100644
index fa84a4f..0000000
--- a/tests/testthat/_snaps/plot_marginal_histograms/mod-cat-pred.svg
+++ /dev/null
@@ -1,76 +0,0 @@
-
-