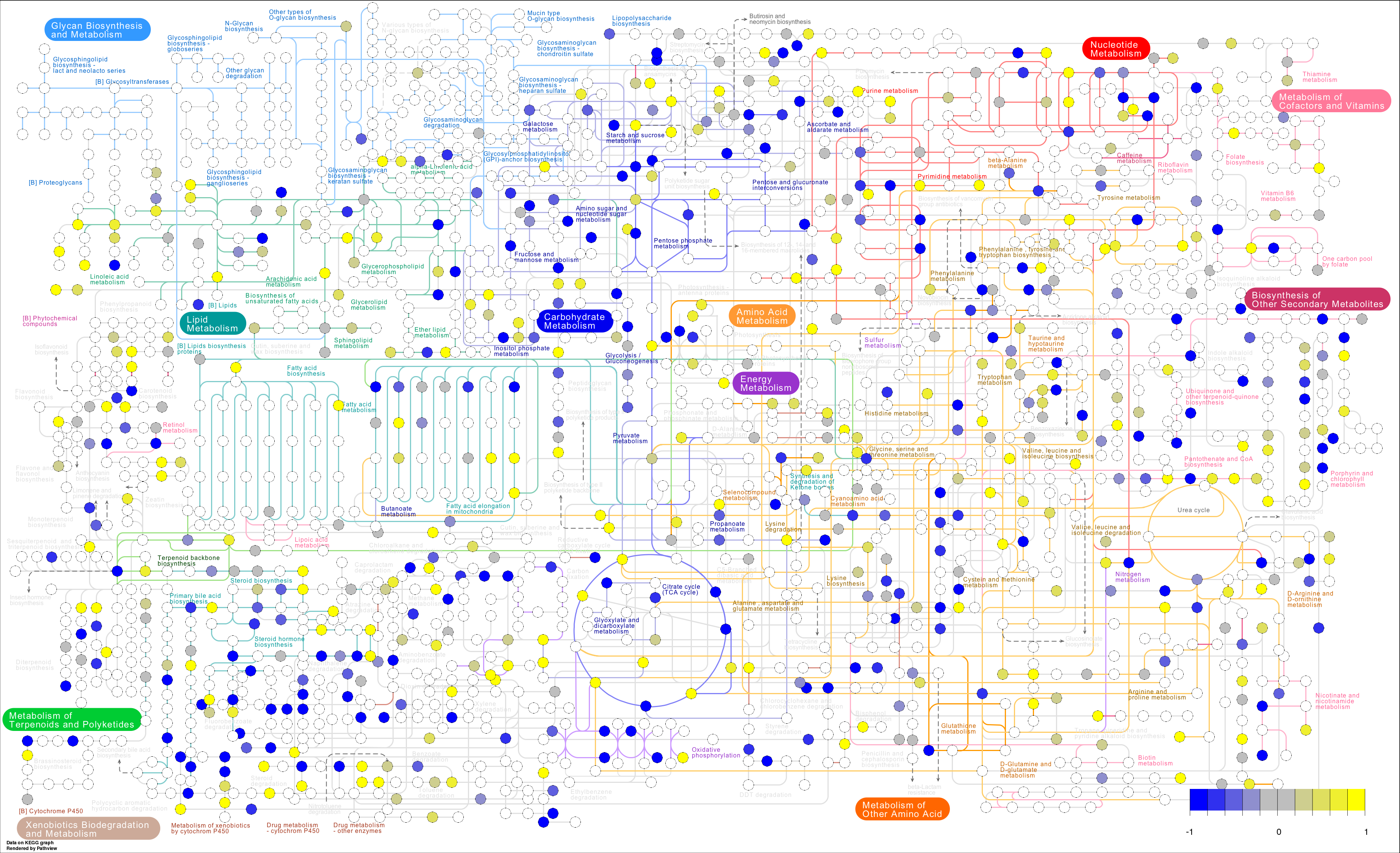
To generate a pathway enrichment visualization we minimally need a list of fold changes in biochemical components of interest (e.g. genes, proteins, metabolites). We need to supply gene/protein and metabolite/compound fold changes separately as data.frames with rownames specifying database identifiers (see pathview for possible options). This can be generated based on the output from statistical analysis used to identify any significantly differentially expressed species.
Lets create gene and metabolite data for a demonstration.
library(pathview)
metabolite.data <- data.frame(FC = sim.mol.data(mol.type = "cpd", nmol = 3000))Lets take a look at the format of the metabolite inputs.
head(metabolite.data)## FC
## C02787 -1.15260
## C08521 0.46416
## C01043 0.72893
## C11496 0.41062
## C07111 -1.46115
## C00031 -0.01891
Notice we have a single column data frame containing log fold changes with KEGG identifiers for the rownames. Next lets load the example gene data. For genes we can specify the organism of interest using the species argument. We can check for available organisms using the commands below.
data(korg)
head(korg)## kegg.code scientific.name common.name
## [1,] "hsa" "Homo sapiens" "human"
## [2,] "ptr" "Pan troglodytes" "chimpanzee"
## [3,] "pps" "Pan paniscus" "bonobo"
## [4,] "ggo" "Gorilla gorilla gorilla" "western lowland gorilla"
## [5,] "pon" "Pongo abelii" "Sumatran orangutan"
## [6,] "mcc" "Macaca mulatta" "rhesus monkey"
## entrez.gnodes kegg.geneid ncbi.geneid
## [1,] "1" "100" "100"
## [2,] "1" "100533953" "100533953"
## [3,] "1" "100967419" "100967419"
## [4,] "1" "101123859" "101123859"
## [5,] "1" "100169736" "100169736"
## [6,] "1" "100301991" "100301991"
We can use the columns scientific.name or common.name to search for the kegg.code for our organism of interest. Here is an example of how we can do this for arabidopsis thaliana.
organism <- "arabidopsis thaliana"
matches <- unlist(sapply(1:ncol(korg), function(i) {
agrep(organism, korg[, i])
}))
(kegg.code <- korg[matches, 1, drop = F])## kegg.code
## [1,] "ath"
# load gene data
gene.data <- data.frame(FC = sim.mol.data(mol.type = "gene", nmol = 3000, species = kegg.code))Lets take a look at the gene data.
head(gene.data)## FC
## AT2G21190 -1.15260
## AT1G80990 0.46416
## AT3G48550 0.72893
## AT1G15410 0.41062
## AT3G15220 -1.46115
## AT3G18980 -0.01891
Looking at the rownames we can see that the identifiers are specific for arabidopsis thaliana. Using real data it is possible that we will need to translate identifiers to match the type used in pathview. Here are the different identifiers which can be supplied for metabolites.
data(cpd.simtypes)
cpd.simtypes## [1] "Beilstein Registry Number" "CAS Registry Number"
## [3] "ChEMBL COMPOUND" "KEGG COMPOUND accession"
## [5] "KEGG DRUG accession" "Patent accession"
## [7] "PubMed citation"
Here are some possible options for gene identifiers.
data(gene.idtype.list)
gene.idtype.list## [1] "SYMBOL" "GENENAME" "ENSEMBL" "ENSEMBLPROT"
## [5] "PROSITE" "UNIGENE" "UNIPROT" "ACCNUM"
## [9] "ENSEMBLTRANS" "REFSEQ"
We can optionally simulate data with with other identifiers by selecting the appropriate identifier type for the argument id.type in the function sim.mol.data .
In the example data above we have the the logarithm of fold changes for 3000 metabolites and 3000 genes. Now that we have the data we can select some pathway of interest to map the fold changes to. We could get this information from a pathway enrichment analysis. We would do this by testing if the significantly differential expressed metabolites or genes are enriched for some specific KEGG pathways of interest. Here are some tools to conduct enrichment analysis for genes and metabolites.
- MBrole (metabolites)
- MetaboAnalyst (metabolites)
- David (genes)
- IMPaLA (genes and metabolites)
For the example below we will randomly select some KEGG pathways.
We can use the R package KEGGREST to get all KEGG pathway identifiers for arabidopsis thaliana.
# get names of pathways to visualize
library(KEGGREST)
pathways <- keggList("pathway", kegg.code)
head(pathways)## path:ath00010
## "Glycolysis / Gluconeogenesis - Arabidopsis thaliana (thale cress)"
## path:ath00020
## "Citrate cycle (TCA cycle) - Arabidopsis thaliana (thale cress)"
## path:ath00030
## "Pentose phosphate pathway - Arabidopsis thaliana (thale cress)"
## path:ath00040
## "Pentose and glucuronate interconversions - Arabidopsis thaliana (thale cress)"
## path:ath00051
## "Fructose and mannose metabolism - Arabidopsis thaliana (thale cress)"
## path:ath00052
## "Galactose metabolism - Arabidopsis thaliana (thale cress)"
Lets visualize changes in our genes and metabolites for Citrate cycle (TCA cycle) - Arabidopsis thaliana (thale cress) which has the KEGG id path:ath00020 .
library(pathview)
map <- gsub("path:", "", names(pathways)[2]) # remove 'path:'
pv.out <- pathview(gene.data = gene.data, cpd.data = metabolite.data, gene.idtype = "KEGG",
pathway.id = map, species = kegg.code, out.suffix = map, keys.align = "y",
kegg.native = T, match.data = T, key.pos = "topright")
plot.name <- paste(map, map, "png", sep = ".")If everything went as planned this generated a file named ath00020.ath00020.png mapped KEGG pathway.
 We can take a look at the mappings made to this pathway.
We can take a look at the mappings made to this pathway.
head(pv.out)## $plot.data.gene
## kegg.names labels type x y width height FC mol.col
## 29 AT1G48030 mtLPD1 gene 467 618 46 17 NA #FFFFFF
## 30 AT3G55410 AT3G55410 gene 661 574 46 17 NA #FFFFFF
## 31 AT3G55410 AT3G55410 gene 530 575 46 17 NA #FFFFFF
## 32 AT4G26910 AT4G26910 gene 403 574 46 17 NA #FFFFFF
## 33 AT2G20420 AT2G20420 gene 260 574 46 17 NA #FFFFFF
## 34 AT2G20420 AT2G20420 gene 260 555 46 17 NA #FFFFFF
## 35 AT1G54340 ICDH gene 718 505 46 17 NA #FFFFFF
## 36 AT5G03290 IDH-IV gene 766 458 46 17 -0.4385 #5FDF5F
## 37 AT1G54340 ICDH gene 718 400 46 17 NA #FFFFFF
## 38 AT1G10670 ACLA-3 gene 434 355 46 17 -1.1941 #00FF00
## 39 AT2G47510 FUM1 gene 191 436 46 17 NA #FFFFFF
## 40 AT2G05710 ACO3 gene 670 345 46 17 NA #FFFFFF
## 41 AT2G05710 ACO3 gene 571 345 46 17 NA #FFFFFF
## 42 AT2G42790 CSY3 gene 434 334 46 17 NA #FFFFFF
## 43 AT1G04410 AT1G04410 gene 253 344 46 17 NA #FFFFFF
## 77 AT1G30120 PDH-E1_ALPHA gene 686 241 46 17 0.6117 #EF3030
## 79 AT1G30120 PDH-E1_ALPHA gene 582 241 46 17 0.6117 #EF3030
## 82 AT1G48030 mtLPD1 gene 529 285 46 17 NA #FFFFFF
## 84 AT1G34430 EMB3003 gene 464 242 46 17 NA #FFFFFF
## 91 AT4G37870 PCK1 gene 371 138 46 17 NA #FFFFFF
## 97 AT5G66760 SDH1-2 gene 191 530 46 17 1.1329 #FF0000
##
## $plot.data.cpd
## kegg.names labels type x y width height FC mol.col
## 58 C00022 C00022 compound 737 241 8 8 0.21082 #CECE8F
## 59 C00122 C00122 compound 190 481 8 8 NA #FFFFFF
## 60 C00036 C00036 compound 320 344 8 8 1.42760 #FFFF00
## 61 C05379 C05379 compound 718 453 8 8 0.76991 #EFEF30
## 62 C00024 C00024 compound 400 241 8 8 NA #FFFFFF
## 63 C00149 C00149 compound 190 393 8 8 1.93744 #FFFF00
## 64 C00311 C00311 compound 718 344 8 8 -1.33400 #0000FF
## 65 C00417 C00417 compound 621 344 8 8 -0.82811 #0000FF
## 66 C00042 C00042 compound 190 574 8 8 1.28575 #FFFF00
## 67 C00158 C00158 compound 522 344 8 8 NA #FFFFFF
## 68 C15972 C15972 compound 522 617 8 8 NA #FFFFFF
## 69 C00068 C00068 compound 589 530 8 8 0.01167 #BEBEBE
## 70 C16254 C16254 compound 462 573 8 8 NA #FFFFFF
## 71 C15973 C15973 compound 408 617 8 8 NA #FFFFFF
## 72 C00091 C00091 compound 334 574 8 8 -1.97370 #0000FF
## 73 C00026 C00026 compound 718 573 8 8 1.10432 #FFFF00
## 74 C05381 C05381 compound 593 573 8 8 NA #FFFFFF
## 76 C05125 C05125 compound 631 241 8 8 NA #FFFFFF
## 78 C00068 C00068 compound 631 198 8 8 0.01167 #BEBEBE
## 80 C15972 C15972 compound 580 284 8 8 NA #FFFFFF
## 83 C16255 C16255 compound 517 241 8 8 NA #FFFFFF
## 93 C15973 C15973 compound 478 284 8 8 NA #FFFFFF
## 94 C00074 C00074 compound 471 127 8 8 NA #FFFFFF
We can also display changes in genes/proteins and metabolites for multiple comparisons. To do this lets create some more artificial data and this time lets get protein information.
metabolite.data2 <- sim.mol.data(mol.type = "cpd", nmol = 3000, nexp = 2)
head(metabolite.data2)## exp1 exp2
## C02787 -1.15260 1.2914
## C08521 0.46416 0.6706
## C01043 0.72893 -0.3065
## C11496 0.41062 -2.1996
## C07111 -1.46115 -0.3476
## C00031 -0.01891 0.7219
gene.data2 <- sim.mol.data(mol.type = "gene", nmol = 3000, nexp = 2, id.type = "UNIPROT")
head(gene.data2)## exp1 exp2
## E9PDR7 -1.15260 1.2914
## Q9UBV2 0.46416 0.6706
## Q9BQ95 0.72893 -0.3065
## P24310 0.41062 -2.1996
## Q8N5I0 -1.46115 -0.3476
## Q9Y6J0 -0.01891 0.7219
Notice UNIPROT IDs do not map to arabidopsis thaliana. We can check the available mappings by looking at the database of identifiers for arabidopsis thaliana org.At.tair.db. Lets instead map changes to a human pathway or KEGG code "hsa".
To do this we need to make sure we supply the correct species and gene.idtype arguments. Next lets map changes in proteins and metabolites for the two comparisons to the pathway .
map <- gsub("path:ath", "", names(pathways)[1]) # remove 'path:ath'
pv.out <- pathview(gene.data = gene.data2, cpd.data = metabolite.data2, gene.idtype = "UNIPROT",
pathway.id = map, species = "hsa", out.suffix = map, keys.align = "y", kegg.native = T,
match.data = T, key.pos = "topright")
plot.name <- paste(map, map, "multi", "png", sep = ".")This should have generated an image file named 00010.00010.multi.png.

We could also map metabolites on a large scale using pathway.id= '01100'.
sessionInfo()## R version 3.0.1 (2013-05-16)
## Platform: i386-w64-mingw32/i386 (32-bit)
##
## locale:
## [1] LC_COLLATE=English_United States.1252
## [2] LC_CTYPE=English_United States.1252
## [3] LC_MONETARY=English_United States.1252
## [4] LC_NUMERIC=C
## [5] LC_TIME=English_United States.1252
##
## attached base packages:
## [1] parallel stats graphics grDevices utils datasets methods
## [8] base
##
## other attached packages:
## [1] KEGGREST_1.2.2 pathview_1.2.4 org.Hs.eg.db_2.9.0
## [4] RSQLite_0.11.3 DBI_0.2-7 AnnotationDbi_1.24.0
## [7] Biobase_2.22.0 BiocGenerics_0.8.0 KEGGgraph_1.16.0
## [10] graph_1.40.1 XML_3.96-1.1 knitr_1.5
## [13] rcom_2.2-5 rscproxy_2.0-5
##
## loaded via a namespace (and not attached):
## [1] Biostrings_2.30.1 evaluate_0.5.3 formatR_0.10
## [4] grid_3.0.1 httr_0.3 IRanges_1.20.7
## [7] png_0.1-7 RCurl_1.95-4.1 Rgraphviz_2.6.0
## [10] stats4_3.0.1 stringr_0.6.2 tools_3.0.1
## [13] XVector_0.2.0
