diff --git a/docs/index.md b/docs/index.md
index 0c03410..e3d1549 100644
--- a/docs/index.md
+++ b/docs/index.md
@@ -14,7 +14,7 @@ navigation: 1
-# 
+# 
# Nanopore analysis pipeline v1.5
Nextflow pipeline for analysis of Nanopore data from direct RNA sequencing. This is a joint project between [CRG bioinformatics core](https://biocore.crg.eu/) and [Epitranscriptomics and RNA Dynamics research group](https://www.crg.eu/en/programmes-groups/novoa-lab).
diff --git a/docs/nanomod.md b/docs/nanomod.md
index 668f910..b986048 100644
--- a/docs/nanomod.md
+++ b/docs/nanomod.md
@@ -9,7 +9,7 @@ This module allows to predict the loci with RNA modifications starting from data
## Workflow
-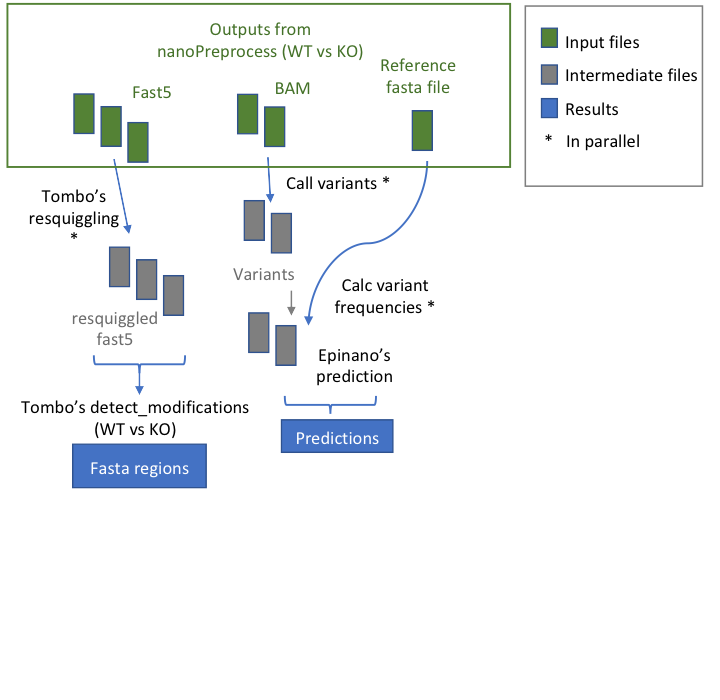 +
+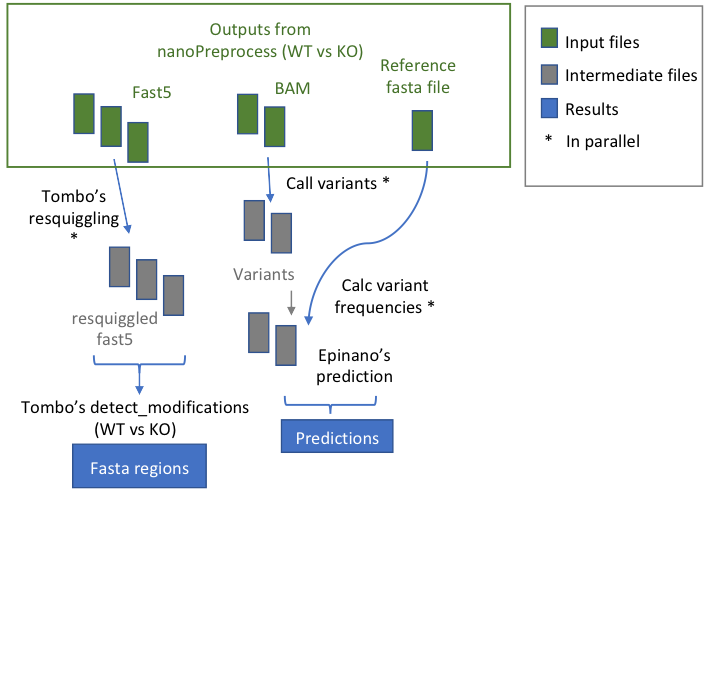 * **index_reference** index the reference file for Epinano
diff --git a/docs/nanopreprocess.md b/docs/nanopreprocess.md
index 7411957..6248afa 100644
--- a/docs/nanopreprocess.md
+++ b/docs/nanopreprocess.md
@@ -11,7 +11,7 @@ This module takes as input the raw fast5 reads - single or multi - and produces
## Workflow
-
* **index_reference** index the reference file for Epinano
diff --git a/docs/nanopreprocess.md b/docs/nanopreprocess.md
index 7411957..6248afa 100644
--- a/docs/nanopreprocess.md
+++ b/docs/nanopreprocess.md
@@ -11,7 +11,7 @@ This module takes as input the raw fast5 reads - single or multi - and produces
## Workflow
-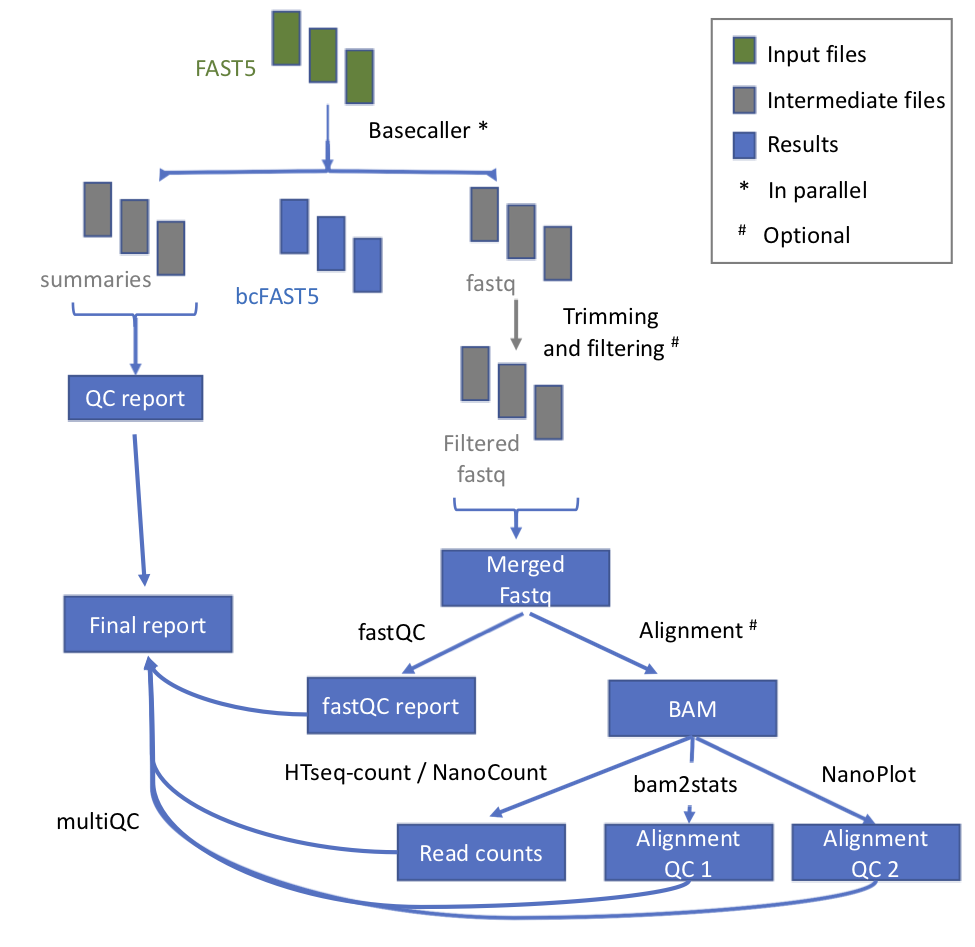 +
+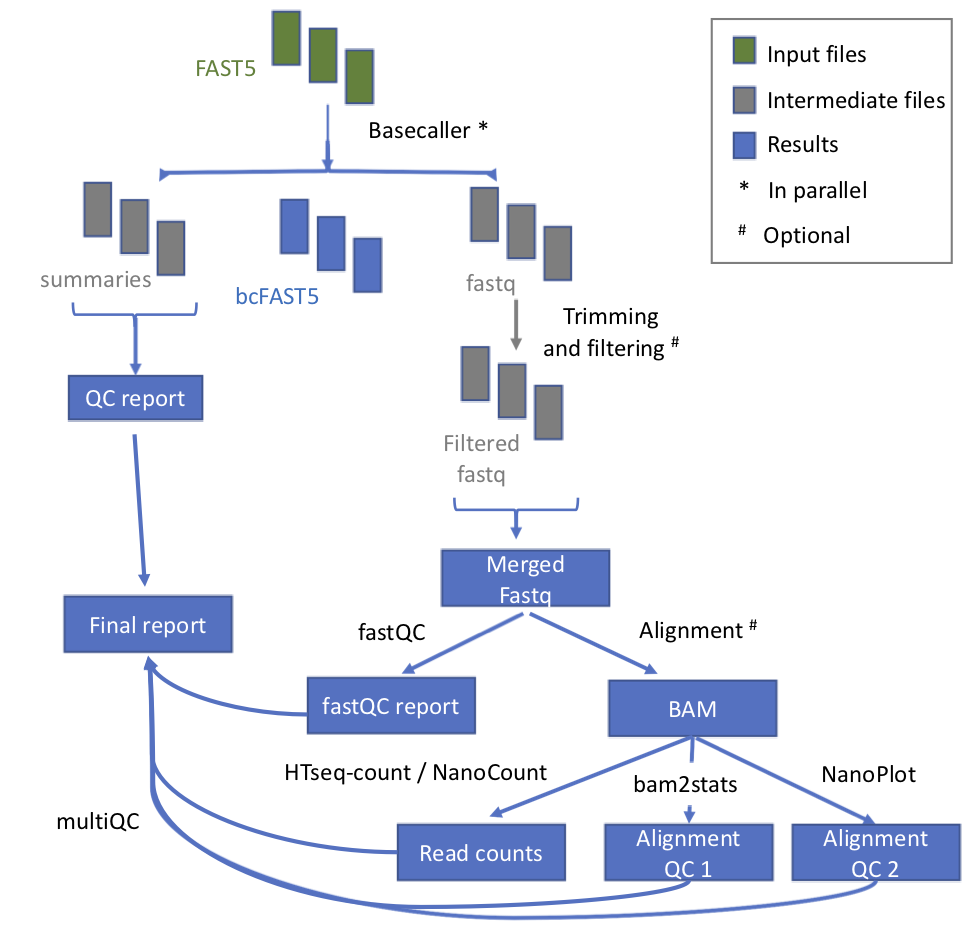 | Process name | Description |
| ------------- | ------------- |
diff --git a/docs/nanotail.md b/docs/nanotail.md
index d49f204..0d9d181 100644
--- a/docs/nanotail.md
+++ b/docs/nanotail.md
@@ -9,7 +9,7 @@ This module allows to estimates polyA sizes by using two different methods (nano
# Workflow
-
| Process name | Description |
| ------------- | ------------- |
diff --git a/docs/nanotail.md b/docs/nanotail.md
index d49f204..0d9d181 100644
--- a/docs/nanotail.md
+++ b/docs/nanotail.md
@@ -9,7 +9,7 @@ This module allows to estimates polyA sizes by using two different methods (nano
# Workflow
-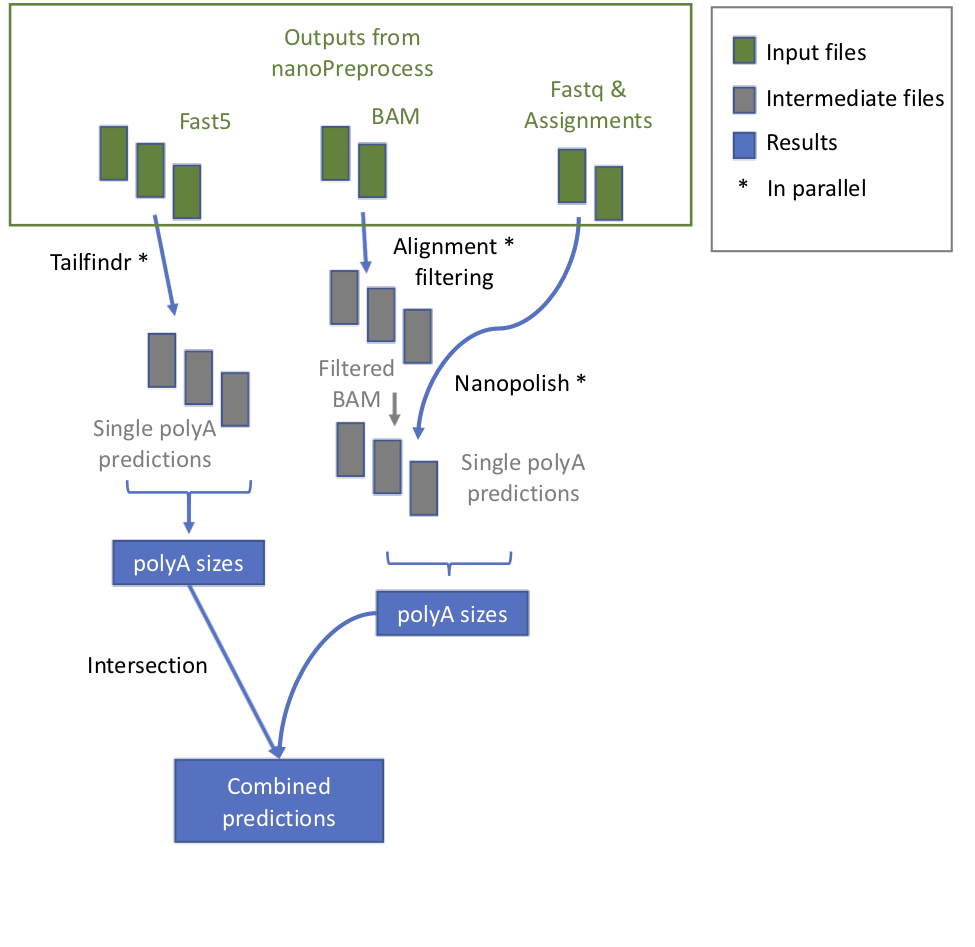 +
+ 1. **check_reference** It verifies whether the reference is zipped and eventually unzip it
1. **tailfindr** it runs *tailfindr* tool in parallel.
1. **check_reference** It verifies whether the reference is zipped and eventually unzip it
1. **tailfindr** it runs *tailfindr* tool in parallel.
 +
+ * **index_reference** index the reference file for Epinano
diff --git a/docs/nanopreprocess.md b/docs/nanopreprocess.md
index 7411957..6248afa 100644
--- a/docs/nanopreprocess.md
+++ b/docs/nanopreprocess.md
@@ -11,7 +11,7 @@ This module takes as input the raw fast5 reads - single or multi - and produces
## Workflow
-
* **index_reference** index the reference file for Epinano
diff --git a/docs/nanopreprocess.md b/docs/nanopreprocess.md
index 7411957..6248afa 100644
--- a/docs/nanopreprocess.md
+++ b/docs/nanopreprocess.md
@@ -11,7 +11,7 @@ This module takes as input the raw fast5 reads - single or multi - and produces
## Workflow
- +
+ | Process name | Description |
| ------------- | ------------- |
diff --git a/docs/nanotail.md b/docs/nanotail.md
index d49f204..0d9d181 100644
--- a/docs/nanotail.md
+++ b/docs/nanotail.md
@@ -9,7 +9,7 @@ This module allows to estimates polyA sizes by using two different methods (nano
# Workflow
-
| Process name | Description |
| ------------- | ------------- |
diff --git a/docs/nanotail.md b/docs/nanotail.md
index d49f204..0d9d181 100644
--- a/docs/nanotail.md
+++ b/docs/nanotail.md
@@ -9,7 +9,7 @@ This module allows to estimates polyA sizes by using two different methods (nano
# Workflow
- +
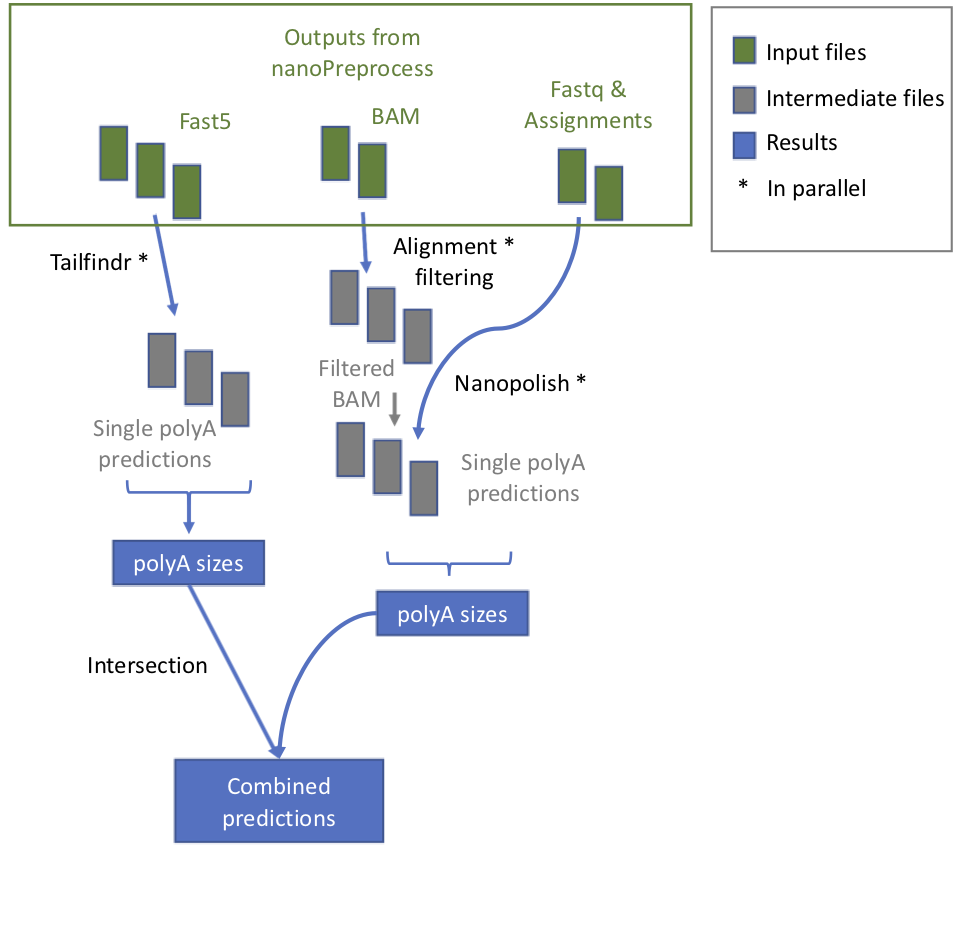
+ 1. **check_reference** It verifies whether the reference is zipped and eventually unzip it
1. **tailfindr** it runs *tailfindr* tool in parallel.
1. **check_reference** It verifies whether the reference is zipped and eventually unzip it
1. **tailfindr** it runs *tailfindr* tool in parallel.