diff --git a/docs/tutorial_group_fdr.md b/docs/tutorial_group_fdr.md
new file mode 100644
index 000000000..6f2ec8eff
--- /dev/null
+++ b/docs/tutorial_group_fdr.md
@@ -0,0 +1,33 @@
+## Perform group FDR estimation for different identification groups
+
+##### [FragPipe](https://fragpipe.nesvilab.org) can be downloaded [here](https://github.com/Nesvilab/FragPipe/releases). Follow the instructions on that same Releases page to launch the program. See [here](https://fragpipe.nesvilab.org/docs/tutorial_fragpipe.html#configure-fragpipe) for help configuring FragPipe.
+
+FragPipe can perform a group FDR estimation to calculate FDRs for each group separately. Then groups can be the number of enzymatic termini (_i.e._ 0, 1, 2) and the protein evidence levels encoded in the FASTA file.
+
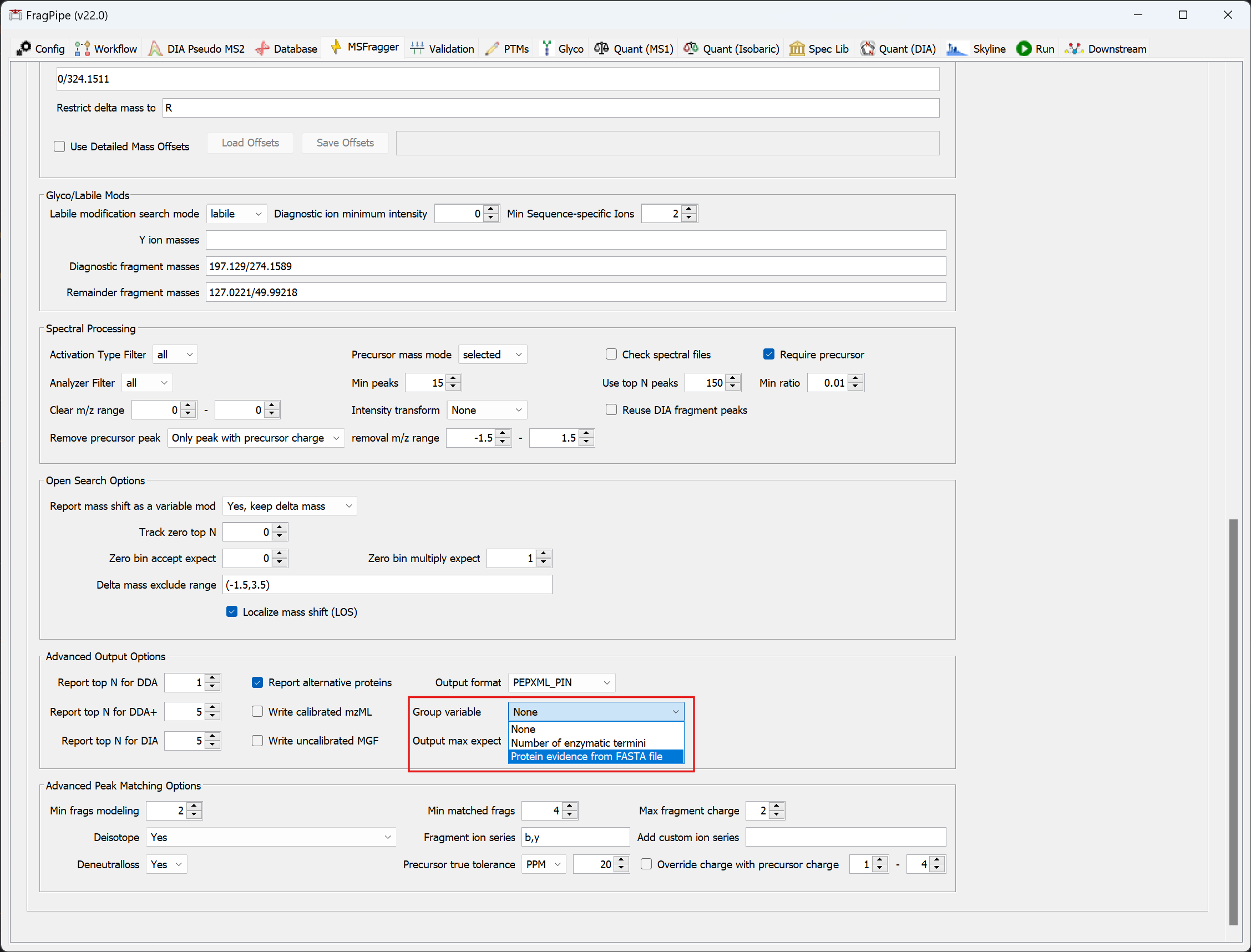
+The group FDR approach supports most workflows. After loading a specific workflow, go to the MSFragger tab to select the group type.
+
+
+
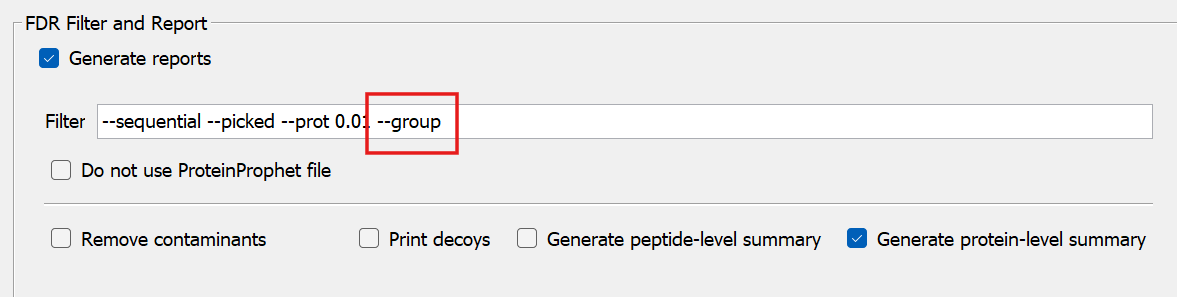
+Then, go to the `Validation` tab, and add `--group` to the `FDR Filter and Report` command box
+
+
+
+The rest of the part is the same as that in a normal FragPipe analysis.
+
+To make FragPipe recognize the encoded protein evidence, the protein header in the FASTA file must have the `PE=` keyword. The numbers after `PE=` identify different groups. [Here](https://github.com/bassanilab/CircRNA_MS_ref_fasta) has a handy script to revise the protein headers.
+
+
+
+
+
+
+
+#### Key References
+_coming soon_
+
+
+
+
+
+
+#### [Back to FragPipe homepage](https://fragpipe.nesvilab.org/)