diff --git a/README.md b/README.md
index 07ac319..755497a 100644
--- a/README.md
+++ b/README.md
@@ -6,7 +6,7 @@ The **GrIML** processing package for classifying water bodies from satellite ima
## Installation
-The GrIML package can be installed using pip:
+The GrIML post-processing Python package can be installed using pip:
```
$ pip install griml
@@ -21,16 +21,17 @@ $ pip install .
## Workflow outline
-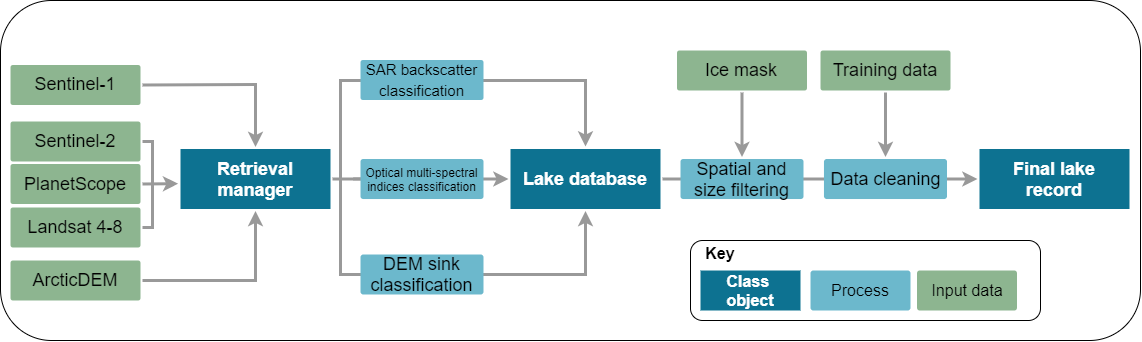 +
+ **GrIML** proposes to examine ice marginal lake changes across Greenland using a multi-sensor and multi-method remote sensing approach to better address their influence on sea level contribution forecasting.
-Ice marginal lakes are detected using a remote sensing approach, based on offline workflows developed within the [ESA Glaciers CCI](https://catalogue.ceda.ac.uk/uuid/7ea7540135f441369716ef867d217519") (Option 6, An Inventory of Ice-Marginal Lakes in Greenland) ([How et al., 2021](https://www.nature.com/articles/s41598-021-83509-1)). Lake extents are defined through a multi-sensor approach using:
+Ice marginal lakes are detected using a remote sensing approach, based on offline workflows developed within the [ESA Glaciers CCI](https://catalogue.ceda.ac.uk/uuid/7ea7540135f441369716ef867d217519") (Option 6, An Inventory of Ice-Marginal Lakes in Greenland) ([How et al., 2021](https://www.nature.com/articles/s41598-021-83509-1)). Initial classifications are performed on Google Earth Engine with the scripts available [here](https://github.com/PennyHow/GrIML/tree/main/gee_scripts). Lake extents are defined through a multi-sensor approach using:
- Multi-spectral indices classification from Sentinel-2 optical imagery
- Backscatter classification from Sentinel-1 SAR (synthetic aperture radar) imagery
- Sink detection from ArcticDEM digital elevation models
+Post-processing of these classifications is performed using the GrIML post-processing Python package, including raster-to-vector conversion, filtering, merging, metadata population, and statistical analysis.
## Project links
diff --git a/src/griml.egg-info/PKG-INFO b/src/griml.egg-info/PKG-INFO
deleted file mode 100644
index f83ffdc..0000000
--- a/src/griml.egg-info/PKG-INFO
+++ /dev/null
@@ -1,76 +0,0 @@
-Metadata-Version: 2.1
-Name: griml
-Version: 0.0.1
-Summary: A workflow for classifying lakes from satellite imagery and compiling lake inventories
-Home-page: https://github.com/PennyHow/GrIML
-Author: Penelope How
-Author-email: pho@geus.dk
-License: UNKNOWN
-Project-URL: Bug Tracker, https://github.com/PennyHow/GrIML/issues
-Keywords: glaciology ice lake ESA
-Platform: UNKNOWN
-Classifier: Programming Language :: Python :: 3
-Classifier: License :: OSI Approved :: MIT License
-Classifier: Development Status :: 4 - Beta
-Classifier: Intended Audience :: Science/Research
-Classifier: Natural Language :: English
-Classifier: Topic :: Scientific/Engineering
-Classifier: Operating System :: OS Independent
-Requires-Python: >=3.6
-Description-Content-Type: text/markdown
-License-File: LICENSE
-
-# Investigating Greenland's ice marginal lakes under a changing climate (GrIML)
-
-[](https://griml.readthedocs.io/en/latest/?badge=latest)
-
-A repository for all project-related materials, funded under the ESA Living Planet Fellowship.
-
-**Project aim:** To examine ice marginal lake changes across Greenland using a multi-method and multi-sensor remote sensing approach, refined with in situ validation.
-
-## Background
-
-
**GrIML** proposes to examine ice marginal lake changes across Greenland using a multi-sensor and multi-method remote sensing approach to better address their influence on sea level contribution forecasting.
-Ice marginal lakes are detected using a remote sensing approach, based on offline workflows developed within the [ESA Glaciers CCI](https://catalogue.ceda.ac.uk/uuid/7ea7540135f441369716ef867d217519") (Option 6, An Inventory of Ice-Marginal Lakes in Greenland) ([How et al., 2021](https://www.nature.com/articles/s41598-021-83509-1)). Lake extents are defined through a multi-sensor approach using:
+Ice marginal lakes are detected using a remote sensing approach, based on offline workflows developed within the [ESA Glaciers CCI](https://catalogue.ceda.ac.uk/uuid/7ea7540135f441369716ef867d217519") (Option 6, An Inventory of Ice-Marginal Lakes in Greenland) ([How et al., 2021](https://www.nature.com/articles/s41598-021-83509-1)). Initial classifications are performed on Google Earth Engine with the scripts available [here](https://github.com/PennyHow/GrIML/tree/main/gee_scripts). Lake extents are defined through a multi-sensor approach using:
- Multi-spectral indices classification from Sentinel-2 optical imagery
- Backscatter classification from Sentinel-1 SAR (synthetic aperture radar) imagery
- Sink detection from ArcticDEM digital elevation models
+Post-processing of these classifications is performed using the GrIML post-processing Python package, including raster-to-vector conversion, filtering, merging, metadata population, and statistical analysis.
## Project links
diff --git a/src/griml.egg-info/PKG-INFO b/src/griml.egg-info/PKG-INFO
deleted file mode 100644
index f83ffdc..0000000
--- a/src/griml.egg-info/PKG-INFO
+++ /dev/null
@@ -1,76 +0,0 @@
-Metadata-Version: 2.1
-Name: griml
-Version: 0.0.1
-Summary: A workflow for classifying lakes from satellite imagery and compiling lake inventories
-Home-page: https://github.com/PennyHow/GrIML
-Author: Penelope How
-Author-email: pho@geus.dk
-License: UNKNOWN
-Project-URL: Bug Tracker, https://github.com/PennyHow/GrIML/issues
-Keywords: glaciology ice lake ESA
-Platform: UNKNOWN
-Classifier: Programming Language :: Python :: 3
-Classifier: License :: OSI Approved :: MIT License
-Classifier: Development Status :: 4 - Beta
-Classifier: Intended Audience :: Science/Research
-Classifier: Natural Language :: English
-Classifier: Topic :: Scientific/Engineering
-Classifier: Operating System :: OS Independent
-Requires-Python: >=3.6
-Description-Content-Type: text/markdown
-License-File: LICENSE
-
-# Investigating Greenland's ice marginal lakes under a changing climate (GrIML)
-
-[](https://griml.readthedocs.io/en/latest/?badge=latest)
-
-A repository for all project-related materials, funded under the ESA Living Planet Fellowship.
-
-**Project aim:** To examine ice marginal lake changes across Greenland using a multi-method and multi-sensor remote sensing approach, refined with in situ validation.
-
-## Background
-
- -
-Sea level is predicted to rise drastically by 2100, with significant contribution from the melting of the Greenland Ice Sheet (GrIS). In these predictions, melt runoff is assumed to contribute directly to sea level change, with little consideration for meltwater storage at the terrestrial margin of the ice sheet; such as ice marginal lakes.
-
-In 2017, 3347 ice marginal lakes were identified in Greenland along the ice margin (How et al., 2021, see map figure for all mapped lakes). Globally, these ice marginal lakes hold up to 0.43 mm of sea level equivalent, which could have a marked impact on future predictions (Shugar et al., 2021). Therefore, they need to be monitored to understand how changes in ice marginal lake water storage affect melt contribution, and how their dynamics evolve under a changing climate.
-
-**GrIML** proposes to examine ice marginal lake changes across Greenland using a multi-sensor and multi-method remote sensing approach to better address their influence on sea level contribution forecasting.
-
-1. Greenland-wide inventories of ice marginal lakes will be generated for selected years during the satellite era, building upon established classification methods in a unified cloud processing workflow
-
-2. Detailed time-series analysis will be conducted on chosen ice marginal lakes to assess changes in their flooding dynamics; focusing on lakes with societal and scientific importance
-
-3. The findings from this work will be validated against in situ observations - namely hydrological measurements and terrestrial time-lapse images - to evaluate whether the remote sensing workflow adequately captures ice marginal lake dynamics
-
-
-## Methodology
-
-Ice marginal lakes will be detected using a remote sensing approach, based on offline workflows developed within the ESA Glaciers CCI (Option 6, An Inventory of Ice-Marginal Lakes in Greenland) (see workflow below). Lake extents were defined through a multi-sensor approach, using multi-spectral indices classification from Sentinel-2 optical imagery, backscatter classification from Sentinel-1 SAR (synthetic aperture radar) imagery, and sink detection from ArcticDEM digital elevation models (How et al., 2021).
-
-
-
-Sea level is predicted to rise drastically by 2100, with significant contribution from the melting of the Greenland Ice Sheet (GrIS). In these predictions, melt runoff is assumed to contribute directly to sea level change, with little consideration for meltwater storage at the terrestrial margin of the ice sheet; such as ice marginal lakes.
-
-In 2017, 3347 ice marginal lakes were identified in Greenland along the ice margin (How et al., 2021, see map figure for all mapped lakes). Globally, these ice marginal lakes hold up to 0.43 mm of sea level equivalent, which could have a marked impact on future predictions (Shugar et al., 2021). Therefore, they need to be monitored to understand how changes in ice marginal lake water storage affect melt contribution, and how their dynamics evolve under a changing climate.
-
-**GrIML** proposes to examine ice marginal lake changes across Greenland using a multi-sensor and multi-method remote sensing approach to better address their influence on sea level contribution forecasting.
-
-1. Greenland-wide inventories of ice marginal lakes will be generated for selected years during the satellite era, building upon established classification methods in a unified cloud processing workflow
-
-2. Detailed time-series analysis will be conducted on chosen ice marginal lakes to assess changes in their flooding dynamics; focusing on lakes with societal and scientific importance
-
-3. The findings from this work will be validated against in situ observations - namely hydrological measurements and terrestrial time-lapse images - to evaluate whether the remote sensing workflow adequately captures ice marginal lake dynamics
-
-
-## Methodology
-
-Ice marginal lakes will be detected using a remote sensing approach, based on offline workflows developed within the ESA Glaciers CCI (Option 6, An Inventory of Ice-Marginal Lakes in Greenland) (see workflow below). Lake extents were defined through a multi-sensor approach, using multi-spectral indices classification from Sentinel-2 optical imagery, backscatter classification from Sentinel-1 SAR (synthetic aperture radar) imagery, and sink detection from ArcticDEM digital elevation models (How et al., 2021).
-
- -
-The intent in GrIML is to build upon this pre-existing workflow with new and innovative solutions to form a unified and automated processing chain, with the option to integrate it into a cloud processing platform for efficient big data analysis.
-
-These developments will alleviate the current challenges associated with data-heavy processing (i.e. multi-sensor integration and data retrieval), and ensure detection accuracy with a merged and collated set of established methodologies. These developments will include:
-
-1. Incorporation of the multi-spectral indices classification, backscatter classification, and sink detection methods into one unified processing chain
-
-2. Integration of image analysis from additional sensors into the processing chain, such as Landsat 4-8
-
-3. Automisation of post-processing filtering to remove detached water bodies and misclassifications, using the already-existing 2017 inventory (How et al., 2021) as a training dataset to automatically match and retain existing lakes
-
-4. Adaptation of the workflow for easy transference between offline and cloud processing platforms
-
-## Links
-
-- ESA project outline and fellow information
-
-- Information about the ESA Living Planet Fellowship
-
-- GrIML project description
-
-- 2017 ice marginal lake inventory Scientific Reports paper and dataset
-
-
diff --git a/src/griml.egg-info/SOURCES.txt b/src/griml.egg-info/SOURCES.txt
deleted file mode 100644
index b38c4c2..0000000
--- a/src/griml.egg-info/SOURCES.txt
+++ /dev/null
@@ -1,15 +0,0 @@
-LICENSE
-README.md
-setup.py
-src/griml/__init__.py
-src/griml/dem.py
-src/griml/lake.py
-src/griml/process.py
-src/griml/retrieve.py
-src/griml/sar.py
-src/griml/vis.py
-src/griml.egg-info/PKG-INFO
-src/griml.egg-info/SOURCES.txt
-src/griml.egg-info/dependency_links.txt
-src/griml.egg-info/requires.txt
-src/griml.egg-info/top_level.txt
\ No newline at end of file
diff --git a/src/griml.egg-info/dependency_links.txt b/src/griml.egg-info/dependency_links.txt
deleted file mode 100644
index 8b13789..0000000
--- a/src/griml.egg-info/dependency_links.txt
+++ /dev/null
@@ -1 +0,0 @@
-
diff --git a/src/griml.egg-info/requires.txt b/src/griml.egg-info/requires.txt
deleted file mode 100644
index fe00572..0000000
--- a/src/griml.egg-info/requires.txt
+++ /dev/null
@@ -1,6 +0,0 @@
-earthengine-api
-geopandas
-numpy
-pandas
-scipy
-Shapely
diff --git a/src/griml.egg-info/top_level.txt b/src/griml.egg-info/top_level.txt
deleted file mode 100644
index ba4ef6c..0000000
--- a/src/griml.egg-info/top_level.txt
+++ /dev/null
@@ -1 +0,0 @@
-griml
diff --git a/src/griml/examples/getInventory_funcs.py b/src/griml/examples/getInventory_funcs.py
deleted file mode 100644
index 0dea76c..0000000
--- a/src/griml/examples/getInventory_funcs.py
+++ /dev/null
@@ -1,406 +0,0 @@
-"""
-GrIML processing module playground (experimental)
-
-@author: Penelope How
-"""
-
-import ee, time, sys
-from time import gmtime, strftime
-import requests, zipfile, io, urllib
-from urllib.error import HTTPError
-
-import numpy as np
-from shapely.ops import split
-from shapely.geometry import Polygon, LineString, MultiPolygon
-import geopandas as gpd
-import multiprocessing as mp
-
-sys.path.append('../')
-from retrieve import getScenes, getScene, getInt, getSmooth, getMosaic, \
- getMean, maskImage, splitBBox, getVectors, extractFeatures, getFeatures, \
- getFeaturesSplit
-from sar import filterSARscenes, classifySARimage
-from vis import filterS2scenes, maskS2clouds, renameS2scenes, \
- resampleS2scenes, classifyVISimage, filterLSscenes, maskL8clouds, \
- renameL8scenes
-from dem import getElevation, getSlope, getSinks
-from lake import compileFeatures, assignID, assignSources, assignNames
-
-start=time.time()
-
-#------------------------------------------------------------------------------
-
-# Set AOI
-
-aoi1 = [-50.94, 67.98] # SW test region
-aoi2 = [-49.66, 67.58]
-
-# aoi1 = [-54.68242187500001,68.8415979398901]
-# aoi2 = [-47.21171875000001,60.62185574504481] # SW region
-
-# aoi1 = [-10.297656250000014,59.03190534154833] # All Greenland
-# aoi2 = [-74.98515625000002,83.9867103173338]
-
-print('Loading bounding box')
-aoi = '/home/pho/Desktop/python_workspace/GrIML/other/datasets/aoi/test_mask_4326.shp'
-aoi = gpd.read_file(aoi)
-aoi_poly = aoi.geometry.iloc[0]
-xmin, ymin, xmax, ymax = aoi_poly.bounds
-
-# Set AOI box splitter parameters
-wh=0.3 # Window height
-ww=0.3 # Window width
-oh = 0.05 # Overlap height
-ow = 0.05 # Overlap width
-
-# Set date range
-date1='2017-07-01'
-date2='2017-08-31'
-
-# Set maximum cloud cover
-cloud=50
-
-# Set mask location
-ice_mask = 'users/pennyruthhow/GIMP_iceMask_edit_buffer7km'
-
-# Set output projection
-proj = 'EPSG:3413' # Polar stereographic
-
-
-#--------------------------- Initialise GEE -------------------------------
-
-ee.Initialize()
-print('EE initialized')
-
-# Set the Area of Interest (AOI) through box coordinates
-box = ee.Geometry.Rectangle([xmin, ymin, xmax, ymax])
-
-# Split box into grid for exporting
-bbox = splitBBox(aoi_poly, wh, ww, oh, ow)
-print('grid completed')
-# # Remove boxes that don't overlap with AOI polygon
-# grid = [Polygon([[g[0],g[1]], [g[0],g[3]], [g[2],g[3]],
-# [g[2],g[1]], [g[0],g[1]]]) for g in grid]
-# bbox = [ee.Geometry.Rectangle(min(g.exterior.coords.xy[0]),
-# min(g.exterior.coords.xy[1]),
-# max(g.exterior.coords.xy[0]),
-# max(g.exterior.coords.xy[1])) for g in grid if g.intersects(aoi_poly)]
-
-
-print(f'Computed {len(bbox)} bounding boxes')
-print(f'Total area covered: {aoi_poly.area} sq km')
-
-
-
-
-# bbox = bbox[100:105]
-
-# # Set the Area of Interest (AOI) through box coordinates
-# box = ee.Geometry.Rectangle([aoi1[0], aoi1[1],
-# aoi2[0], aoi2[1]])
-
-# bbox = [ee.Geometry.Rectangle(g[0], g[1], g[2], g[3]) for g in grid]
-
-# print(f'Computed {len(bbox)} bounding boxes from {aoi1}, {aoi2}')
-# print(f'Total area covered: {box.area().getInfo()/10**6} sq km')
-
-
-#--------------------- Get basic ocean mask ---------------------------------
-
-# Retrieve ice-ocean mask
-img = ee.Image("OSU/GIMP/2000_ICE_OCEAN_MASK")
-ocean_mask = img.select('ocean_mask').eq(0)
-# ice_mask = img.select('ice_mask').eq(0)
-
-# imask0 = ice_mask.reduceToVectors(geometryType='polygon', labelProperty='v', scale=10, bestEffort=True)
-# def _mapBuffer(feature):
-# return feature.buffer(-10000)
-# imask01 = imask0.map(_mapBuffer)
-# imask0 = imask01.reduceToImage(properties=['v'], reducer=ee.Reducer.mean())
-
-# sys.exit(1)
-
-# # Construct vector features
-# def getFeaturesSplit(image, bbox):
-# features=[]
-# for b in range(len(bbox)):
-# print(f'Fetching vectors from bbox {b}...')
-# v = getVectors(image, 10, bbox[b])
-# features.append(extractFeatures(v))
-# features = [val for sublist in features for val in sublist]
-# print(f'{len(features)} vector features identified')
-# return features
-
-# def getFeatures(image, bbox):
-# features=[]
-# v = getVectors(image, 10, bbox)
-# features = extractFeatures(v)
-# print(f'{len(features)} vector features identified')
-# return features
-
-# def getParallelFeatures1(image, bbox):
-# # Parallel process vector generation
-# pool = mp.Pool(mp.cpu_count())
-# v = [pool.apply(getVectors, args=(image, 10, bbox[b])) for b in range(len(bbox))]
-# pool.close()
-
-# # Parallel process feature extraction
-# pool = mp.Pool(mp.cpu_count())
-# features = pool.map(extractFeatures, [row for row in v])
-# pool.close()
-
-# features = [val for sublist in features for val in sublist]
-# print(f'{len(features)} vector features identified')
-# return features
-
-# def getDownload(v):
-# link = v.getDownloadURL('csv')
-# req = urllib.request.Request(url=link)
-# try:
-# handler = urllib.request.urlopen(req)
-# except HTTPError as e:
-# handler = e.read()
-# lines = []
-# for l in handler:
-# lines.append(str(l))
-# features_dem.append(lines)
-
-#--------------------------- DEM processing -------------------------------
-
-# Set collection
-dem_col = 'UMN/PGC/ArcticDEM/V3/2m_mosaic'
-
-# Get image collection
-scenes = getScene(dem_col, box)
-print(f'\nScenes gathered from {dem_col}')
-
-# Get elevation and slope
-elevation = getElevation(scenes)
-slope = getSlope(elevation)
-
-elevation = getSmooth(elevation, 110)
-
-# Mask out ocean pixels
-elevation = maskImage(elevation, ocean_mask)
-
-# Get sinks
-elevation = getInt(elevation)
-sinks = getSinks(elevation, 10, 50)
-
-# Remove speckle with smoothing
-sinks = getSmooth(sinks, 50).rename('dem_sinks')
-sinks = getInt(sinks)
-print(f'{dem_col} scenes classified')
-print('Band names: ' + str(sinks.bandNames().getInfo()))
-
-try:
- features_dem = getFeatures(sinks, 10, box)
-except:
- features_dem = getFeaturesSplit(sinks, 10, bbox)
-
-
-#--------------------------- SAR processing -------------------------------
-
-# Set collection
-sar_col = 'COPERNICUS/S1_GRD'
-
-# Get image collection
-scenes = getScenes(sar_col, date1, date2, box)
-if scenes.size().getInfo() > 0:
- scenes = filterSARscenes(scenes)
- print(f'\nScenes gathered from {sar_col}')
- print('Total number of scenes: ', scenes.size().getInfo())
- print('Number of bands per scene: '
- + str(len(scenes.first().bandNames().getInfo())))
- print('Band names: ' + str(scenes.first().bandNames().getInfo()))
-
- # Get average
- aver = getMosaic(scenes, 'HH')
-
- # Mask out ocean pixels
- aver = maskImage(aver, ocean_mask)
-
- # Classify water from SAR average image
- water_sar = classifySARimage(aver, -20, 'HH', 50)
- print(f'{sar_col} scenes classified')
- print('Band names: ' + str(water_sar.bandNames().getInfo()))
-
- try:
- features_sar = getFeatures(water_sar, 10, box)
- except:
- features_sar = getFeaturesSplit(water_sar, 10, bbox)
-
-else:
- features_sar = None
- print(f'\nNo {sar_col} scenes identified between {date1} and {date2}')
-
-#------------------------- VIS processing (S2) -----------------------------
-
-# Set collection
-vis_col1 = "COPERNICUS/S2"
-
-# Get image collection
-scenes = getScenes(vis_col1, date1, date2, box)
-if scenes.size().getInfo() > 0:
- scenes = filterS2scenes(scenes, cloud)
- print(f'\nScenes gathered from {vis_col1}')
- print('Total number of scenes: ', scenes.size().getInfo())
- print('Number of bands per scene: '
- + str(len(scenes.first().bandNames().getInfo())))
- print('Band names: ' + str(scenes.first().bandNames().getInfo()))
-
- # Mask scenes for clouds
- scenes = maskS2clouds(scenes)
- # ee.Terrain.hillshade(image, azimuth, elevation)
-
- # Get average of spectific bands
- scenes = renameS2scenes(scenes)
- scenes = resampleS2scenes(scenes)
- aver = getMean(scenes, ['blue','green','red','vnir','swir1_1','swir2_1'])
- print('Scenes resampled and mosiacked')
- print('Band names: ' + str(aver.bandNames().getInfo()))
-
- # Classify water from VIS average image, and mask out ocean pixels
- water_s2 = classifyVISimage(aver)
- water_s2 = maskImage(water_s2, ocean_mask)
- print(f'{vis_col1} scenes classified')
- print('Band names: ' + str(water_s2.bandNames().getInfo()))
-
- try:
- features_s2 = getFeatures(water_s2, 10, box)
- except:
- features_s2 = getFeaturesSplit(water_s2, 10, bbox)
-else:
- features_s2 = None
- print(f'\nNo {vis_col1} scenes identified between {date1} and {date2}')
-
-#----------------------- VIS processing (Landsat 8) -----------------------
-
-# Set collection
-vis_col2 = "LANDSAT/LC08/C01/T1_TOA"
-
-# Get image collection
-scenes = getScenes(vis_col2, date1, date2, box)
-if scenes.size().getInfo() > 0:
- scenes = filterLSscenes(scenes, cloud)
- print(f'\nScenes gathered from {vis_col2}')
- print('Total number of scenes: ', scenes.size().getInfo())
- print('Number of bands per scene: '
- + str(len(scenes.first().bandNames().getInfo())))
- print('Band names: ' + str(scenes.first().bandNames().getInfo()))
-
- # Mask scenes for clouds
- scenes = maskL8clouds(scenes)
- # ee.Terrain.hillshade(image, azimuth, elevation)
-
- # Get average of spectific bands
- scenes = renameL8scenes(scenes)
- aver = getMean(scenes, ['blue','green','red','vnir','swir1','swir2'])
- print('Scenes resampled and mosiacked')
- print('Band names: ' + str(aver.bandNames().getInfo()))
-
- # Classify water from VIS average image, and mask out ocean pixels
- ndwi = aver.normalizedDifference(['green_mean', 'vnir_mean']).rename('ndwi')
- mndwi = aver.normalizedDifference(['green_mean','swir1_mean']).rename('mndwi')
- aweish = aver.expression('BLUE + 2.5 * GREEN - 1.5 * (VNIR + SWIR1) - 0.25 * SWIR2',
- {'BLUE' : aver.select('blue_mean'),
- 'GREEN' : aver.select('green_mean'),
- 'SWIR1' : aver.select('swir1_mean'),
- 'VNIR' : aver.select('vnir_mean'),
- 'SWIR2' : aver.select('swir2_mean')}).rename('aweish')
- aweinsh = aver.expression('4.0 * (GREEN - SWIR1) - (0.25 * VNIR + 2.75 * SWIR2)',
- {'GREEN' : aver.select('green_mean'),
- 'SWIR1' : aver.select('swir1_mean'),
- 'VNIR' : aver.select('vnir_mean'),
- 'SWIR2' : aver.select('swir2_mean')}).rename('aweinsh')
- bright = aver.expression('(RED + GREEN + BLUE) / 3',
- {'BLUE' : aver.select('blue_mean'),
- 'GREEN' : aver.select('green_mean'),
- 'RED' : aver.select('red_mean')}).rename('bright')
-
- aver = aver.addBands([ndwi, mndwi, aweish, aweinsh, bright])
- classified = aver.expression("(BRIGHT > 5000) ? 0"
- ": (NDWI > 0.3) ? 1 "
- ": (MNDWI < 0.1) ? 0 "
- ": (AWEISH < 2000) ? 0"
- ": (AWEISH > 5000) ? 0"
- ": (AWEINSH < 4000) ? 0"
- ": (AWEINSH > 6000) ? 0"
- ": 1",
- {'NDWI' : aver.select('ndwi'),
- 'MNDWI' : aver.select('mndwi'),
- 'AWEISH' : aver.select('aweish'),
- 'AWEINSH' : aver.select('aweinsh'),
- 'BRIGHT' : aver.select('bright')}).rename('water')
- water_ls8 = classified.updateMask(classified)
- water_ls8 = maskImage(water_ls8, ocean_mask)
- print(f'{vis_col2} scenes classified')
- print('Band names: ' + str(water_ls8.bandNames().getInfo()))
-
- try:
- features_ls8 = getFeatures(water_ls8, 10, box)
- except:
- features_ls8 = getFeaturesSplit(water_ls8, 10, bbox)
-
-else:
- features_ls8 = None
- print(f'\nNo {vis_col2} scenes identified between {date1} and {date2}')
-
-
-#------------------------ Compile geodatabase -----------------------------
-
-iml = compileFeatures([features_s2, features_ls8, features_sar, features_dem],
- ['VIS','VIS','SAR','DEM'],
- [vis_col1, vis_col2, sar_col, dem_col],
- [date1, date2])
-print(f'\nCompiled {iml.shape[0]} geodatabase features')
-
-# Reproject geometries
-iml = iml.to_crs(proj)
-print(f'{iml.shape[0]} unfiltered features')
-
-iml.to_file(f"out/iiml_{date1}_{date2}_{aoi1[0]}_{aoi1[1]}_unfiltered.shp")
-
-
-#-------------------- Filter database by ice margin -----------------------
-
-# Load margin and add buffer
-margin = gpd.read_file("../../../other/datasets/ice_margin/gimp_icemask_line_polstereo.shp")
-margin_buff = margin.buffer(500)
-margin_buff = gpd.GeoDataFrame(geometry=margin_buff, crs=margin.crs)
-
-# Perform spatial join
-iml = gpd.sjoin(iml, margin_buff, how='left')
-iml = iml[iml['index_right']==0]
-iml = iml.drop(columns='index_right')
-print(f'{iml.shape[0]} features within 500 m of margin')
-
-# Filter lakes by area
-iml['area_sqkm'] = iml['geometry'].area/10**6
-iml['length_km'] = iml['geometry'].length/1000
-iml = iml[(iml.area_sqkm >= 0.05)]
-
-# Calculate geometry info
-iml.reset_index(inplace=True, drop=True)
-print(f'{iml.shape[0]} features over 0.05 sq km')
-iml.to_file(f"out/iiml_{date1}_{date2}_{aoi1[0]}_{aoi1[1]}_filtered.shp")
-
-#------------------------- Populate metadata ------------------------------
-
-iml = assignID(iml)
-iml = assignSources(iml)
-
-names = gpd.read_file('../../../other/datasets/placenames/oqaasileriffik_placenames.shp')
-iml = assignNames(iml, names)
-
-#----------------------- Write data to shapefile --------------------------
-
-iml.to_file(f"/home/pho/Desktop/python_workspace/GrIML/other/out/iiml_{date1}_{date2}_{aoi1[0]}_{aoi1[1]}_final.shp")
-print(f'Saved to out/iiml_{date1}_{date2}_{aoi1[0]}_{aoi1[1]}_final.shp')
-
-# Plot lakes
-iml.plot(figsize=(10, 10), alpha=0.5, edgecolor='k')
-
-print('\nFinished')
-end=time.time()
-print('Total run time: ' + strftime("%H:%M:%S", gmtime(round(end-start))))
diff --git a/src/griml/examples/getInventory_obj.py b/src/griml/examples/getInventory_obj.py
deleted file mode 100644
index f8b507c..0000000
--- a/src/griml/examples/getInventory_obj.py
+++ /dev/null
@@ -1,182 +0,0 @@
-"""
-GrIML processing module playground (experimental)
-
-@author: Penelope How
-"""
-
-import time,sys
-from time import gmtime, strftime
-import geopandas as gpd
-
-try:
- sys.path.append('../')
- from process import gee
- from lake import dissolvePolygons, compileFeatures, assignID, assignSources, assignNames
-except:
- from griml.process import gee
- from griml.lake import dissolvePolygons, compileFeatures, assignID, \
- assignSources, assignNames
-
-start=time.time()
-
-#------------------------------------------------------------------------------
-
-# Set AOI box splitter parameters
-wh=0.3 # Window height
-ww=0.3 # Window width
-oh = 0.05 # Overlap height
-ow = 0.05 # Overlap width
-
-# Set date range
-date1='2017-07-01'
-date2='2017-08-31'
-
-# Set output projection
-proj = 'EPSG:3413' # Polar stereographic
-
-# Set AOI
-# aoi = '/home/pho/python_workspace/GrIML/other/datasets/aoi/AOI_mask_split2.shp'
-aoi = '/home/pho/python_workspace/GrIML/other/datasets/aoi/test_mask_4326.shp'
-aoi = gpd.read_file(aoi)#.to_crs('EPSG:4326')
-
-# Load ice margin with buffer
-print('Preparing ice margin buffer...')
-margin_buff = gpd.read_file("/home/pho/python_workspace/GrIML/other/datasets/ice_margin/gimp_icemask_line_polstereo_simple_buffer.shp")
-# margin_buff = margin.buffer(500)
-# margin_buff = gpd.GeoDataFrame(geometry=margin_buff, crs=margin.crs)
-
-# Load name database
-print('Loading placename database...')
-names = gpd.read_file('/home/pho/python_workspace/GrIML/other/datasets/placenames/oqaasileriffik_placenames.shp')
-
-
-#--------------------------- Initialise GEE -------------------------------
-
-parameters = [#{'collection' : 'UMN/PGC/ArcticDEM/V3/2m_mosaic',
- #'smooth' : 100, 'fill' : 100, 'kernel' : 100, 'speckle' : 50},
- # {'collection' : 'UMN/PGC/ArcticDEM/V3/2m',
- # 'smooth' : 100, 'fill' : 100, 'kernel' : 100, 'speckle' : 50},
- {'collection' : 'COPERNICUS/S1_GRD',
- 'polar' : 'HH', 'threshold' : -20, 'smooth' : 50},
- #{'collection' : 'COPERNICUS/S2', `
- #'cloud' : 20},
- {'collection' : 'LANDSAT/LC08/C01/T1_TOA',
- 'cloud' : 20}]
- # {'collection' : 'LANDSAT/LE07/C02/T1_TOA',
- # 'cloud' : 50}]
-
-
-
-for i in range(len(aoi.index)):
- print(f'\nCommencing processing of {list(aoi["catch"])[i]} catchment area')
- aoi_poly = aoi.geometry.iloc[i]
- xmin, ymin, xmax, ymax = aoi_poly.bounds
-
- print('Clipping margin to catchment area...')
- aoi_poly_proj = gpd.GeoDataFrame(geometry=[aoi_poly], crs=aoi.crs).to_crs(proj)
- margin_clip = gpd.clip(margin_buff, aoi_poly_proj)
-
- print('Conducting classifications...')
- proc = gee([date1, date2], aoi_poly, parameters, [wh, ww, oh, ow], True)
-
- print('Processing...')
- water = proc.processAll()
-
- print('Retrieving...')
- features = proc.retrieveAll(water)
-
- # Filter features
- filtered=[]
- for f in features:
- print('Filtering features...')
-
- # Remove duplicates
- print('Dissolving polygons...')
- gdf = gpd.GeoDataFrame(geometry=f, crs='EPSG:4326')
- gdf = dissolvePolygons(gdf)
-
- # Remove lakes below size threshold
- print('Filtering by size...')
- gdf = gdf.set_crs('EPSG:4326')
- gdf = gdf.to_crs(proj)
- gdf['area_sqkm'] = gdf['geometry'].area/10**6
- gdf = gdf[(gdf.area_sqkm >= 0.05)]
-
- # # Remove lakes outside of margin boundary
- # print('Filtering by bounds...')
- # gdf = gpd.sjoin(gdf, margin_clip, how='left')
- # gdf = gdf[gdf['index_right']==0]
- # gdf = gdf.drop(columns='index_right')
- filtered.append(gdf.geometry)
-
-
-#------------------------ Compile geodatabase -----------------------------
-# lakes=[]
-# for f in features:
-# gdf = gpd.GeoDataFrame(geometry=f, crs='EPSG:4326')
-# gdf = dissolvePolygons(gdf)
-# gdf['area'] = gdf['geometry'].area/10**6
-# gdf = gdf[(gdf.area >= 0.05)]
-
-# gdf = gpd.sjoin(gdf, margin_buff, how='left')
-# gdf = gdf[gdf['index_right']==0]
-# gdf = gdf.drop(columns='index_right')
-# lakes.append(list(gdf['geometry']))
-
- # Compile all features
- iml = compileFeatures(filtered,
- ['DEM', 'SAR', 'VIS'],
- ['UMN/PGC/ArcticDEM/V3/2m_mosaic', 'COPERNICUS/S1_GRD',
- 'LANDSAT/LC08/C01/T1_TOA'],
- [date1, date2], proj)
- print(f'\nCompiled {iml.shape[0]} geodatabase features')
-
- # Calculate geometry attributes
- iml['area_sqkm'] = iml['geometry'].area/10**6
- iml['length_km'] = iml['geometry'].length/1000
- iml = iml[(iml.area_sqkm >= 0.05)]
- iml['catch'] = str(list(aoi["catch"])[i])
-
- # Calculate geometry info
- iml.reset_index(inplace=True, drop=True)
-
- print(f'{iml.shape[0]} filtered features')
-
- iml.to_file(f'out/iiml_{date1}_{date2}_{list(aoi["catch"])[i]}_filtered.shp')
-
-
-# #-------------------- Filter database by ice margin -----------------------
-
-# # Perform spatial join
-# aoi_poly_proj = aoi_poly.to_crs(proj)
-# margin_clip = gpd.clip(margin_buff, aoi_poly_proj)
-
-# iml = gpd.sjoin(iml, margin_clip, how='left')
-# iml = iml[iml['index_right']==0]
-# iml = iml.drop(columns='index_right')
-# print(f'{iml.shape[0]} features within 500 m of margin')
-
-# # Calculate geometry info
-# iml.reset_index(inplace=True, drop=True)
-# print(f'{iml.shape[0]} features over 0.05 sq km')
-# iml.to_file(f'out/iiml_{date1}_{date2}_{list(aoi["catch"])[i]}_filtered.shp')
-
-
-#-------------------- Populate metadata and write to file -----------------
-
-# iml = assignID(iml)
- # iml = assignSources(iml)
-
- # iml = assignNames(iml, names)
-
- # iml.to_file(f'out/iiml_{date1}_{date2}_{list(aoi["catch"])[i]}_final.shp')
- # print(f'Saved to out/iiml_{date1}_{date2}_{list(aoi["catch"])[i]}_final.shp')
-
- # # Plot lakes
- # iml.plot(figsize=(10, 10), alpha=0.5, edgecolor='k')
-
-
-#------------------------------------------------------------------------------
-print('\nFinished')
-end=time.time()
-print('Total run time: ' + strftime("%H:%M:%S", gmtime(round(end-start))))
diff --git a/src/griml/metadata/__init__.py b/src/griml/metadata/__init__.py
index 3d16982..7e1e3d8 100644
--- a/src/griml/metadata/__init__.py
+++ b/src/griml/metadata/__init__.py
@@ -1,5 +1,6 @@
from griml.metadata.assign_certainty import *
from griml.metadata.assign_id import *
from griml.metadata.assign_names import *
+from griml.metadata.assign_regions import *
from griml.metadata.assign_sources import *
from griml.metadata.add_metadata import *
diff --git a/src/griml/metadata/add_metadata.py b/src/griml/metadata/add_metadata.py
index 2f15ee2..50e6d9a 100644
--- a/src/griml/metadata/add_metadata.py
+++ b/src/griml/metadata/add_metadata.py
@@ -1,30 +1,17 @@
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
-from griml.metadata import assign_id, assign_sources, assign_certainty, \
- assign_names
from griml.load import load
+from griml.metadata import assign_id, assign_sources, assign_certainty, \
+ assign_names, assign_regions
import geopandas as gpd
-def add_metadata(iml_file, names_file, outfile=None):
- '''Add metadata to collection of features
-
- Parameters
- ----------
- iml_file : str, geopandas.dataframe.DataFrame
- Filepath or geopandas.dataframe.DataFrame object to assign metadata to
-
- Returns
- -------
- iml: geopandas.dataframe.GeoDataFrame
- Geodataframe with metadata
- '''
-
- # Load files
- iml = load(iml_file)
- names = load(names_file)
+def add_metadata(iml, names, regions, outfile=None):
+
+ iml = load(iml)
+ names = load(names)
+ regions = load(regions)
- # Perform metadata steps
print('Assigning ID...')
iml = assign_id(iml)
@@ -35,24 +22,27 @@ def add_metadata(iml_file, names_file, outfile=None):
n = ['S1','S2','ARCTICDEM']
scores = [0.298, 0.398, 0.304]
iml = assign_certainty(iml, n, scores)
+
+ print('Assigning regions...')
+ iml = assign_regions(iml, regions)
print('Assigning placenames...')
iml = assign_names(iml, names)
- # Save to file if given
- if outfile is not None:
+ if outfile:
print('Saving file...')
iml.to_file(outfile)
- print('Saved to '+str(outfile))
-
- return iml
+ print('Saved to '+str(outfile)+"_metadata.shp")
if __name__ == "__main__":
- infile1 = '../test/test_merge_2.shp'
- infile2 = '../test/test_placenames.shp'
- add_metadata(infile1, infile2)
+ # indir = "/home/pho/python_workspace/GrIML/other/iml_2017/merged_vectors/griml_2017_inventory.shp"
+ indir = "/home/pho/python_workspace/GrIML/other/iml_2017/inspected_vectors/lakes_all-0000000000-0000037888_filtered.shp"
+
+
+ infile_names = '/home/pho/python_workspace/GrIML/other/datasets/placenames/oqaasileriffik_placenames.shp'
+ infile_regions = '/home/pho/python_workspace/GrIML/other/datasets/drainage_basins/greenland_basins_polarstereo.shp'
+
- iml = gpd.read_file(infile1)
- names = gpd.read_file(infile2)
- add_metadata(iml, names)
\ No newline at end of file
+ outfile = "/home/pho/python_workspace/GrIML/other/iml_2017/metadata_vectors/griml_2017_inventory_final.shp"
+ add_metadata(indir, infile_names, infile_regions, outfile)
diff --git a/src/griml/metadata/assign_names.py b/src/griml/metadata/assign_names.py
index 9bd294f..c151a48 100644
--- a/src/griml/metadata/assign_names.py
+++ b/src/griml/metadata/assign_names.py
@@ -1,7 +1,18 @@
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
-def assign_names(gdf, gdf_names, distance=500.0):
+import itertools
+from operator import itemgetter
+
+import geopandas as gpd
+import numpy as np
+import pandas as pd
+
+from scipy.spatial import cKDTree
+from shapely.geometry import Point, LineString, Polygon
+from griml.load import load
+
+def assign_names(gdf, gdf_names):
'''Assign placenames to geodataframe geometries based on names in another
geodataframe point geometries
@@ -19,62 +30,41 @@ def assign_names(gdf, gdf_names, distance=500.0):
gdf : pandas.GeoDataFrame
Vectors with assigned IDs
'''
- placenames = _compile_names(gdf_names)
+
+ # Load geodataframes
+ gdf1 = load(gdf)
+ gdf2 = load(gdf_names)
+
+ # Compile placenames into new dataframe
+ names = _compile_names(gdf2)
+ placenames = gpd.GeoDataFrame({"geometry": list(gdf2['geometry']),
+ "placename": names})
- lakename=[]
- for i,v in gdf.iterrows():
-
- geom = v['geometry']
-
- polynames=[]
- for pt in range(len(placenames)):
- if geom.contains(gdf_names['geometry'][pt]) == True:
- polynames.append(placenames[pt])
-
- if len(polynames)==0:
- for pt in range(len(placenames)):
- if gdf_names['geometry'][pt].distance(geom) < distance:
- polynames.append(placenames[pt])
- lakename.append(polynames)
-
- elif len(polynames)==1:
- lakename.append(polynames)
-
- else:
- out=[]
- for p in polynames:
- out.append(p)
- lakename.append(out)
-
- lakeid = gdf['unique_id'].tolist()
- lakename2 = []
+ # Assign names based on proximity
+ a = _get_nearest_point(gdf1, placenames)
+ return a
+
+
+def _get_nearest_point(gdA, gdB, distance=500.0):
+ '''Return properties of nearest point in Y to geometry in X'''
+ nA = np.array(list(gdA.geometry.centroid.apply(lambda x: (x.x, x.y))))
+ nB = np.array(list(gdB.geometry.apply(lambda x: (x.x, x.y))))
+
+ btree = cKDTree(nB)
+ dist, idx = btree.query(nA, k=1)
+ gdB_nearest = gdB.iloc[idx].drop(columns="geometry").reset_index(drop=True)
+ gdf = pd.concat(
+ [
+ gdA.reset_index(drop=True),
+ gdB_nearest,
+ pd.Series(dist, name='dist')
+ ],
+ axis=1)
- for x in gdf.index:
- indx = _get_indices(lakeid, x)
- findname=[]
- for l in indx:
- if len(lakename[l])!=0:
- findname.append(lakename[l])
-
- for i in range(len(indx)):
- if len(findname)==0:
- lakename2.append('')
-
- else:
- unique = set(findname[0])
- unique = list(unique)
-
- if len(unique)==1:
- lakename2.append(findname[0][0])
-
- else:
- out2 = ', '
- out2 = out2.join(unique)
- lakename2.append(out2)
- gdf['placename'] = lakename2
+ gdf.loc[gdf['dist']>=distance, 'placename'] = 'Unknown'
+ gdf = gdf.drop(columns=['dist'])
return gdf
-
def _get_indices(mylist, value):
'''Get indices for value in list'''
return[i for i, x in enumerate(mylist) if x==value]
@@ -95,3 +85,14 @@ def _compile_names(gdf):
else:
placenames.append(None)
return placenames
+
+
+if __name__ == "__main__":
+
+ # Define file attributes
+ infile = '/home/pho/Desktop/python_workspace/GrIML/src/griml/test/test_merge_1.shp'
+ names = '/home/pho/Desktop/python_workspace/GrIML/other/datasets/placenames/oqaasileriffik_placenames_polarstereo.shp'
+
+
+ # Perform conversion
+ a = assign_names(infile, names)
\ No newline at end of file
diff --git a/src/griml/metadata/assign_region.py b/src/griml/metadata/assign_region.py
deleted file mode 100644
index 9bd294f..0000000
--- a/src/griml/metadata/assign_region.py
+++ /dev/null
@@ -1,97 +0,0 @@
-#!/usr/bin/env python3
-# -*- coding: utf-8 -*-
-
-def assign_names(gdf, gdf_names, distance=500.0):
- '''Assign placenames to geodataframe geometries based on names in another
- geodataframe point geometries
-
- Parameters
- ----------
- gdf : pandas.GeoDataFrame
- Vectors to assign uncertainty to
- gdf_names : pandas.GeoDataFrame
- Vector geodataframe with placenames
- distance : int
- Distance threshold between a given vector and a placename
-
- Returns
- -------
- gdf : pandas.GeoDataFrame
- Vectors with assigned IDs
- '''
- placenames = _compile_names(gdf_names)
-
- lakename=[]
- for i,v in gdf.iterrows():
-
- geom = v['geometry']
-
- polynames=[]
- for pt in range(len(placenames)):
- if geom.contains(gdf_names['geometry'][pt]) == True:
- polynames.append(placenames[pt])
-
- if len(polynames)==0:
- for pt in range(len(placenames)):
- if gdf_names['geometry'][pt].distance(geom) < distance:
- polynames.append(placenames[pt])
- lakename.append(polynames)
-
- elif len(polynames)==1:
- lakename.append(polynames)
-
- else:
- out=[]
- for p in polynames:
- out.append(p)
- lakename.append(out)
-
- lakeid = gdf['unique_id'].tolist()
- lakename2 = []
-
- for x in gdf.index:
- indx = _get_indices(lakeid, x)
- findname=[]
- for l in indx:
- if len(lakename[l])!=0:
- findname.append(lakename[l])
-
- for i in range(len(indx)):
- if len(findname)==0:
- lakename2.append('')
-
- else:
- unique = set(findname[0])
- unique = list(unique)
-
- if len(unique)==1:
- lakename2.append(findname[0][0])
-
- else:
- out2 = ', '
- out2 = out2.join(unique)
- lakename2.append(out2)
- gdf['placename'] = lakename2
- return gdf
-
-
-def _get_indices(mylist, value):
- '''Get indices for value in list'''
- return[i for i, x in enumerate(mylist) if x==value]
-
-
-def _compile_names(gdf):
- '''Get preferred placenames from placename geodatabase'''
- placenames=[]
- for i,v in gdf.iterrows():
- if v['Ny_grønla'] != None:
- placenames.append(v['Ny_grønla'])
- else:
- if v['Dansk'] != None:
- placenames.append(v['Dansk'])
- else:
- if v['Alternativ'] != None:
- placenames.append(v['Alternativ'])
- else:
- placenames.append(None)
- return placenames
diff --git a/src/griml/metadata/assign_regions.py b/src/griml/metadata/assign_regions.py
new file mode 100644
index 0000000..6458989
--- /dev/null
+++ b/src/griml/metadata/assign_regions.py
@@ -0,0 +1,86 @@
+#!/usr/bin/env python3
+# -*- coding: utf-8 -*-
+
+import itertools
+from operator import itemgetter
+
+import geopandas as gpd
+import numpy as np
+import pandas as pd
+
+from griml.load import load
+from scipy.spatial import cKDTree
+from shapely.geometry import Point, LineString, Polygon
+
+def assign_regions(gdf, gdf_regions, region_name='region'):
+ '''Assign region to geodataframe geometries based on regions in another
+ geodataframe object
+
+ Parameters
+ ----------
+ gdf : pandas.GeoDataFrame
+ Vectors to assign region name to
+ gdf_regions : pandas.GeoDataFrame
+ Vector geodataframe with regions
+
+ Returns
+ -------
+ gdf : pandas.GeoDataFrame
+ Vectors with assigned IDs
+ '''
+
+ # Load geodataframes
+ gdf1 = load(gdf)
+ gdf2 = load(gdf_regions)
+
+ g = _get_nearest_polygon(gdf1, gdf2)
+
+ return g
+
+
+def _get_nearest_polygon(gdfA, gdfB, gdfB_cols=['subregion'], distance=100000.0):
+ '''Return given properties of nearest polygon in Y to geometry in X'''
+
+ A = np.array(list(gdfA.geometry.centroid.apply(lambda x: (x.x, x.y))))
+ # B = [np.array(geom.boundary.coords) for geom in gdfB.geometry.to_list()]
+
+ B=[]
+ for geom in gdfB.geometry.to_list():
+ try:
+ g = np.array(geom.boundary.coords)
+ except:
+ g = np.array(geom.boundary.geoms[0].coords)
+ B.append(g)
+
+
+ B_ix = tuple(itertools.chain.from_iterable(
+ [itertools.repeat(i, x) for i, x in enumerate(list(map(len, B)))]))
+ B = np.concatenate(B)
+ ckd_tree = cKDTree(B)
+
+ dist, idx = ckd_tree.query(A, k=1)
+ idx = itemgetter(*idx)(B_ix)
+
+ gdf = pd.concat(
+ [gdfA, gdfB.loc[idx, gdfB_cols].reset_index(drop=True),
+ pd.Series(dist, name='dist')], axis=1)
+
+ gdf.loc[gdf['dist']>=distance, 'subregion'] = 'Unknown'
+
+ gdf = gdf.drop(columns=['dist'])
+ return gdf
+
+
+if __name__ == "__main__":
+
+ # Define file attributes
+ infile = '/home/pho/Desktop/python_workspace/GrIML/src/griml/test/test_merge_1.shp'
+ regions = '/home/pho/python_workspace/GrIML/other/datasets/drainage_basins/greenland_basins_polarstereo.shp'
+ regions=gpd.read_file(regions)
+
+ regions_dissolve = regions.dissolve(by='subregion')
+
+
+ # Perform conversion
+ a = assign_regions(infile, regions)
+
diff --git a/src/griml/metadata/metadata.py b/src/griml/metadata/metadata.py
deleted file mode 100644
index 9b0ca30..0000000
--- a/src/griml/metadata/metadata.py
+++ /dev/null
@@ -1,38 +0,0 @@
-#!/usr/bin/env python3
-# -*- coding: utf-8 -*-
-
-from griml.metadata import assign_id, assign_sources, assign_certainty, \
- assign_names
-import geopandas as gpd
-
-def add_metadata(iml, names, outfile):
-
- print('Assigning ID...')
- iml = assign_id(iml)
-
- print('Assigning sources...')
- iml = assign_sources(iml)
-
- print('Assigning certainty scores...')
- n = ['S1','S2','ARCTICDEM']
- scores = [0.298, 0.398, 0.304]
- iml = assign_certainty(iml, n, scores)
-
- print('Assigning placenames...')
- iml = assign_names(iml, names)
-
- print('Saving file...')
- iml.to_file(outfile)
- print('Saved to '+str(outfile)+"_metadata.shp")
-
-
-if __name__ == "__main__":
- # indir = "/home/pho/python_workspace/GrIML/other/iml_2017/merged_vectors/griml_2017_inventory.shp"
- indir = "/home/pho/python_workspace/GrIML/other/iml_2017/inspected_vectors/lakes_all-0000000000-0000037888_filtered.shp"
- iml = gpd.read_file(indir)
-
- infile_names = '/home/pho/python_workspace/GrIML/other/datasets/placenames/oqaasileriffik_placenames.shp'
- names = gpd.read_file(infile_names)
-
- outfile = "/home/pho/python_workspace/GrIML/other/iml_2017/metadata_vectors/griml_2017_inventory_final.shp"
- add_metadata(iml, names, outfile)
diff --git a/src/griml/test/greenland_basins_polarstereo.cpg b/src/griml/test/greenland_basins_polarstereo.cpg
new file mode 100644
index 0000000..cd89cb9
--- /dev/null
+++ b/src/griml/test/greenland_basins_polarstereo.cpg
@@ -0,0 +1 @@
+ISO-8859-1
\ No newline at end of file
diff --git a/src/griml/test/greenland_basins_polarstereo.dbf b/src/griml/test/greenland_basins_polarstereo.dbf
new file mode 100644
index 0000000..9d97cd3
Binary files /dev/null and b/src/griml/test/greenland_basins_polarstereo.dbf differ
diff --git a/src/griml/test/greenland_basins_polarstereo.prj b/src/griml/test/greenland_basins_polarstereo.prj
new file mode 100644
index 0000000..2e49174
--- /dev/null
+++ b/src/griml/test/greenland_basins_polarstereo.prj
@@ -0,0 +1 @@
+PROJCS["WGS_1984_NSIDC_Sea_Ice_Polar_Stereographic_North",GEOGCS["GCS_WGS_1984",DATUM["D_WGS_1984",SPHEROID["WGS_1984",6378137.0,298.257223563]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Stereographic_North_Pole"],PARAMETER["False_Easting",0.0],PARAMETER["False_Northing",0.0],PARAMETER["Central_Meridian",-45.0],PARAMETER["Standard_Parallel_1",70.0],UNIT["Meter",1.0]]
\ No newline at end of file
diff --git a/src/griml/test/greenland_basins_polarstereo.shp b/src/griml/test/greenland_basins_polarstereo.shp
new file mode 100644
index 0000000..2773592
Binary files /dev/null and b/src/griml/test/greenland_basins_polarstereo.shp differ
diff --git a/src/griml/test/greenland_basins_polarstereo.shx b/src/griml/test/greenland_basins_polarstereo.shx
new file mode 100644
index 0000000..a63669b
Binary files /dev/null and b/src/griml/test/greenland_basins_polarstereo.shx differ
diff --git a/src/griml/test/process_cli.py b/src/griml/test/process_cli.py
new file mode 100644
index 0000000..915effc
--- /dev/null
+++ b/src/griml/test/process_cli.py
@@ -0,0 +1,132 @@
+#!/usr/bin/env python3
+# -*- coding: utf-8 -*-
+"""
+@author: pho
+"""
+from griml.convert import convert
+from griml.filter import filter_vectors
+from griml.merge import merge_vectors
+from griml.metadata import add_metadata
+from pathlib import Path
+import glob, os
+from argparse import ArgumentParser
+
+def parse_griml_arguments():
+ parser = ArgumentParser(description="Full post-processing workflow for "+
+ "creating ice marginal lake inventory")
+ parser.add_argument('-r', '--root_dir', type=str, required=True,
+ help='Root directory to write files to')
+ parser.add_argument('-i', '--in_dir', type=str, required=True,
+ help='Directory path to input raster files')
+ parser.add_argument('-y', '--year', type=str, required=True,
+ help='Year of inventory')
+ parser.add_argument('-m', '--margin_file', type=str, required=True,
+ help='File path to ice margin for spatial filtering')
+ parser.add_argument('-n', '--names_file', type=str, required=True,
+ help='File path to placenames file for metadata population')
+ parser.add_argument('-p', '--proj', type=str, default='EPSG:3413',
+ required=False, help='Projection (of input and output)')
+ parser.add_argument('-s', '--steps', type=str, default='1111',
+ required=False, help='Define which steps to include in'+
+ ' processing, where each value indicates: convert, '+
+ 'filter, merge, and metadata. If set to zero, the '+
+ 'step associated with that position is skipped')
+
+ args = parser.parse_args()
+ return args
+
+def get_step_flags(a):
+ '''Return step flags'''
+ return a[0],a[1],a[2],a[3]
+
+
+def check_dir(d):
+ '''Check if directory exists and create it if it does not'''
+ if not os.path.exists(d):
+ os.mkdir(d)
+
+
+def griml_process():
+ '''Perform processing workflow'''
+ args = parse_griml_arguments()
+
+ s1, s2, s3, s4 = get_step_flags(args.steps)
+
+ print('Commencing post processing for inventory year ' + args.year)
+ print('Adopted projection: ' + args.proj)
+ print('Writing outputs to ' + args.root_dir)
+
+ root_dir = Path(args.root_dir)
+
+ # Convert to vectors
+ if s1:
+ print('Converting rasters to vectors...')
+
+ src=args.in_dir
+ dest = str(root_dir.joinpath('vectors'))
+ check_dir(dest)
+
+ print('Reading from ' + src)
+ print('Writing to ' + dest)
+
+ band_info = [{'b_number':1, 'method':'VIS', 'source':'S2'},
+ {'b_number':2, 'method':'SAR', 'source':'S1'},
+ {'b_number':3, 'method':'DEM', 'source':'ARCTICDEM'}]
+ start=args.year+'0701'
+ end=args.year+'0831'
+
+ infiles = list(glob.glob(src+'/*.tif'))
+
+ convert(infiles, args.proj, band_info, start, end, str(dest))
+
+
+ # Filter vectors by area and proximity to margin
+ if s2:
+ print('Filtering vectors...')
+
+ src = str(root_dir.joinpath('vectors'))
+ dest = str(root_dir.joinpath('filtered'))
+ check_dir(dest)
+
+ print('Reading from ' + src)
+ print('Writing to ' + dest)
+
+ # margin_buff = gpd.read_file(infile_margin)
+ # margin_buff = margin.buffer(500)
+ # margin_buff = gpd.GeoDataFrame(geometry=margin_buff, crs=margin.crs)
+
+ infiles = list(glob.glob(src+'/*.shp'))
+
+ filter_vectors(infiles, args.margin_file, dest)
+
+
+ # Merge vectors
+ if s3:
+ print('Merging vectors...')
+
+ src = str(root_dir.joinpath('filtered'))
+ dest = root_dir.joinpath('merged/'+args.year+'_merged.shp')
+ check_dir(dest.parent)
+
+ print('Reading from ' + src)
+ print('Writing to ' + str(dest))
+
+ infiles = list(glob.glob(src+'/*.shp'))
+
+ merge_vectors(infiles, str(dest))
+
+
+ # Add metadata
+ if s4:
+ print('Adding metadata...')
+
+ src = str(root_dir.joinpath('merged/'+args.year+'_merged.shp'))
+ dest = root_dir.joinpath('metadata/'+args.year+'_metadata.shp')
+ check_dir(dest.parent)
+
+ print('Reading from ' + src)
+ print('Writing to ' + str(dest))
+
+ add_metadata(src, args.names_file, str(dest))
+
+ print('Finished')
diff --git a/src/griml/test/process_example.py b/src/griml/test/process_example.py
new file mode 100644
index 0000000..c96a4d9
--- /dev/null
+++ b/src/griml/test/process_example.py
@@ -0,0 +1,85 @@
+#!/usr/bin/env python3
+# -*- coding: utf-8 -*-
+"""
+@author: pho
+"""
+from griml.convert import convert
+from griml.filter import filter_vectors
+from griml.merge import merge_vectors
+from griml.metadata import add_metadata
+from pathlib import Path
+import glob
+
+root_dir = Path('/home/pho/python_workspace/GrIML/other/')
+year='2016'
+
+print('Commencing post processing for inventory year ' + year)
+
+# Convert to vectors
+print('Converting rasters to vectors...')
+
+src = str(root_dir.joinpath('iml_2016-2023/rasters/'+year+'_iml'))
+dest = str(root_dir.joinpath('iml_2016-2023/vectors/'+year))
+
+print('Reading from ' + src)
+print('Writing to ' + dest)
+
+proj = 'EPSG:3413'
+band_info = [{'b_number':1, 'method':'VIS', 'source':'S2'},
+ {'b_number':2, 'method':'SAR', 'source':'S1'},
+ {'b_number':3, 'method':'DEM', 'source':'ARCTICDEM'}]
+start=year+'0701'
+end=year+'0831'
+
+infiles = list(glob.glob(src+'/*.tif'))
+
+convert(infiles, proj, band_info, start, end, str(dest))
+
+
+# Filter vectors by area and proximity to margin
+print('Filtering vectors...')
+
+src = dest
+dest = str(root_dir.joinpath('iml_2016-2023/filtered/'+year))
+infile_margin = str(root_dir.joinpath('datasets/ice_margin/gimp_icemask_line_polstereo_simple_buffer.shp'))
+
+print('Reading from ' + src)
+print('Writing to ' + dest)
+
+# margin_buff = gpd.read_file(infile_margin)
+# margin_buff = margin.buffer(500)
+# margin_buff = gpd.GeoDataFrame(geometry=margin_buff, crs=margin.crs)
+
+infiles = list(glob.glob(src+'/*.shp'))
+
+filter_vectors(infiles, infile_margin, dest)
+
+
+# Merge vectors
+print('Merging vectors...')
+
+src = dest
+dest = str(root_dir.joinpath('iml_2016-2023/merged/'+year+'_merged.shp'))
+
+print('Reading from ' + src)
+print('Writing to ' + dest)
+
+infiles = list(glob.glob(src+'/*.shp'))
+
+merge_vectors(infiles, dest)
+
+
+# Add metadata
+print('Adding metadata...')
+
+src = dest
+dest = str(root_dir.joinpath('iml_2016-2023/metadata/'+year+'_metadata.shp'))
+
+print('Reading from ' + src)
+print('Writing to ' + dest)
+
+infile_names = str(root_dir.joinpath('datasets/placenames/oqaasileriffik_placenames.shp'))
+
+add_metadata(src, infile_names, dest)
+
+print('Finished')
diff --git a/src/griml/test/test.py b/src/griml/test/test.py
index 336fdee..24346c3 100644
--- a/src/griml/test/test.py
+++ b/src/griml/test/test.py
@@ -17,7 +17,6 @@ def test_convert(self):
{'b_number':3, 'method':'DEM', 'source':'ARCTICDEM'}]
start='20170701'
end='20170831'
-
infile = os.path.join(os.path.dirname(griml.__file__),'test/test_north_greenland.tif')
convert([infile], proj, band_info, start, end)
@@ -37,7 +36,8 @@ def test_metadata(self):
'''Test metadata population'''
infile1 = os.path.join(os.path.dirname(griml.__file__),'test/test_merge_2.shp')
infile2 = os.path.join(os.path.dirname(griml.__file__),'test/test_placenames.shp')
- add_metadata(infile1, infile2)
+ infile3 = os.path.join(os.path.dirname(griml.__file__),'test/greenland_basins_polarstereo.shp')
+ add_metadata(infile1, infile2, infile3)
if __name__ == "__main__":
unittest.main()
-
-The intent in GrIML is to build upon this pre-existing workflow with new and innovative solutions to form a unified and automated processing chain, with the option to integrate it into a cloud processing platform for efficient big data analysis.
-
-These developments will alleviate the current challenges associated with data-heavy processing (i.e. multi-sensor integration and data retrieval), and ensure detection accuracy with a merged and collated set of established methodologies. These developments will include:
-
-1. Incorporation of the multi-spectral indices classification, backscatter classification, and sink detection methods into one unified processing chain
-
-2. Integration of image analysis from additional sensors into the processing chain, such as Landsat 4-8
-
-3. Automisation of post-processing filtering to remove detached water bodies and misclassifications, using the already-existing 2017 inventory (How et al., 2021) as a training dataset to automatically match and retain existing lakes
-
-4. Adaptation of the workflow for easy transference between offline and cloud processing platforms
-
-## Links
-
-- ESA project outline and fellow information
-
-- Information about the ESA Living Planet Fellowship
-
-- GrIML project description
-
-- 2017 ice marginal lake inventory Scientific Reports paper and dataset
-
-
diff --git a/src/griml.egg-info/SOURCES.txt b/src/griml.egg-info/SOURCES.txt
deleted file mode 100644
index b38c4c2..0000000
--- a/src/griml.egg-info/SOURCES.txt
+++ /dev/null
@@ -1,15 +0,0 @@
-LICENSE
-README.md
-setup.py
-src/griml/__init__.py
-src/griml/dem.py
-src/griml/lake.py
-src/griml/process.py
-src/griml/retrieve.py
-src/griml/sar.py
-src/griml/vis.py
-src/griml.egg-info/PKG-INFO
-src/griml.egg-info/SOURCES.txt
-src/griml.egg-info/dependency_links.txt
-src/griml.egg-info/requires.txt
-src/griml.egg-info/top_level.txt
\ No newline at end of file
diff --git a/src/griml.egg-info/dependency_links.txt b/src/griml.egg-info/dependency_links.txt
deleted file mode 100644
index 8b13789..0000000
--- a/src/griml.egg-info/dependency_links.txt
+++ /dev/null
@@ -1 +0,0 @@
-
diff --git a/src/griml.egg-info/requires.txt b/src/griml.egg-info/requires.txt
deleted file mode 100644
index fe00572..0000000
--- a/src/griml.egg-info/requires.txt
+++ /dev/null
@@ -1,6 +0,0 @@
-earthengine-api
-geopandas
-numpy
-pandas
-scipy
-Shapely
diff --git a/src/griml.egg-info/top_level.txt b/src/griml.egg-info/top_level.txt
deleted file mode 100644
index ba4ef6c..0000000
--- a/src/griml.egg-info/top_level.txt
+++ /dev/null
@@ -1 +0,0 @@
-griml
diff --git a/src/griml/examples/getInventory_funcs.py b/src/griml/examples/getInventory_funcs.py
deleted file mode 100644
index 0dea76c..0000000
--- a/src/griml/examples/getInventory_funcs.py
+++ /dev/null
@@ -1,406 +0,0 @@
-"""
-GrIML processing module playground (experimental)
-
-@author: Penelope How
-"""
-
-import ee, time, sys
-from time import gmtime, strftime
-import requests, zipfile, io, urllib
-from urllib.error import HTTPError
-
-import numpy as np
-from shapely.ops import split
-from shapely.geometry import Polygon, LineString, MultiPolygon
-import geopandas as gpd
-import multiprocessing as mp
-
-sys.path.append('../')
-from retrieve import getScenes, getScene, getInt, getSmooth, getMosaic, \
- getMean, maskImage, splitBBox, getVectors, extractFeatures, getFeatures, \
- getFeaturesSplit
-from sar import filterSARscenes, classifySARimage
-from vis import filterS2scenes, maskS2clouds, renameS2scenes, \
- resampleS2scenes, classifyVISimage, filterLSscenes, maskL8clouds, \
- renameL8scenes
-from dem import getElevation, getSlope, getSinks
-from lake import compileFeatures, assignID, assignSources, assignNames
-
-start=time.time()
-
-#------------------------------------------------------------------------------
-
-# Set AOI
-
-aoi1 = [-50.94, 67.98] # SW test region
-aoi2 = [-49.66, 67.58]
-
-# aoi1 = [-54.68242187500001,68.8415979398901]
-# aoi2 = [-47.21171875000001,60.62185574504481] # SW region
-
-# aoi1 = [-10.297656250000014,59.03190534154833] # All Greenland
-# aoi2 = [-74.98515625000002,83.9867103173338]
-
-print('Loading bounding box')
-aoi = '/home/pho/Desktop/python_workspace/GrIML/other/datasets/aoi/test_mask_4326.shp'
-aoi = gpd.read_file(aoi)
-aoi_poly = aoi.geometry.iloc[0]
-xmin, ymin, xmax, ymax = aoi_poly.bounds
-
-# Set AOI box splitter parameters
-wh=0.3 # Window height
-ww=0.3 # Window width
-oh = 0.05 # Overlap height
-ow = 0.05 # Overlap width
-
-# Set date range
-date1='2017-07-01'
-date2='2017-08-31'
-
-# Set maximum cloud cover
-cloud=50
-
-# Set mask location
-ice_mask = 'users/pennyruthhow/GIMP_iceMask_edit_buffer7km'
-
-# Set output projection
-proj = 'EPSG:3413' # Polar stereographic
-
-
-#--------------------------- Initialise GEE -------------------------------
-
-ee.Initialize()
-print('EE initialized')
-
-# Set the Area of Interest (AOI) through box coordinates
-box = ee.Geometry.Rectangle([xmin, ymin, xmax, ymax])
-
-# Split box into grid for exporting
-bbox = splitBBox(aoi_poly, wh, ww, oh, ow)
-print('grid completed')
-# # Remove boxes that don't overlap with AOI polygon
-# grid = [Polygon([[g[0],g[1]], [g[0],g[3]], [g[2],g[3]],
-# [g[2],g[1]], [g[0],g[1]]]) for g in grid]
-# bbox = [ee.Geometry.Rectangle(min(g.exterior.coords.xy[0]),
-# min(g.exterior.coords.xy[1]),
-# max(g.exterior.coords.xy[0]),
-# max(g.exterior.coords.xy[1])) for g in grid if g.intersects(aoi_poly)]
-
-
-print(f'Computed {len(bbox)} bounding boxes')
-print(f'Total area covered: {aoi_poly.area} sq km')
-
-
-
-
-# bbox = bbox[100:105]
-
-# # Set the Area of Interest (AOI) through box coordinates
-# box = ee.Geometry.Rectangle([aoi1[0], aoi1[1],
-# aoi2[0], aoi2[1]])
-
-# bbox = [ee.Geometry.Rectangle(g[0], g[1], g[2], g[3]) for g in grid]
-
-# print(f'Computed {len(bbox)} bounding boxes from {aoi1}, {aoi2}')
-# print(f'Total area covered: {box.area().getInfo()/10**6} sq km')
-
-
-#--------------------- Get basic ocean mask ---------------------------------
-
-# Retrieve ice-ocean mask
-img = ee.Image("OSU/GIMP/2000_ICE_OCEAN_MASK")
-ocean_mask = img.select('ocean_mask').eq(0)
-# ice_mask = img.select('ice_mask').eq(0)
-
-# imask0 = ice_mask.reduceToVectors(geometryType='polygon', labelProperty='v', scale=10, bestEffort=True)
-# def _mapBuffer(feature):
-# return feature.buffer(-10000)
-# imask01 = imask0.map(_mapBuffer)
-# imask0 = imask01.reduceToImage(properties=['v'], reducer=ee.Reducer.mean())
-
-# sys.exit(1)
-
-# # Construct vector features
-# def getFeaturesSplit(image, bbox):
-# features=[]
-# for b in range(len(bbox)):
-# print(f'Fetching vectors from bbox {b}...')
-# v = getVectors(image, 10, bbox[b])
-# features.append(extractFeatures(v))
-# features = [val for sublist in features for val in sublist]
-# print(f'{len(features)} vector features identified')
-# return features
-
-# def getFeatures(image, bbox):
-# features=[]
-# v = getVectors(image, 10, bbox)
-# features = extractFeatures(v)
-# print(f'{len(features)} vector features identified')
-# return features
-
-# def getParallelFeatures1(image, bbox):
-# # Parallel process vector generation
-# pool = mp.Pool(mp.cpu_count())
-# v = [pool.apply(getVectors, args=(image, 10, bbox[b])) for b in range(len(bbox))]
-# pool.close()
-
-# # Parallel process feature extraction
-# pool = mp.Pool(mp.cpu_count())
-# features = pool.map(extractFeatures, [row for row in v])
-# pool.close()
-
-# features = [val for sublist in features for val in sublist]
-# print(f'{len(features)} vector features identified')
-# return features
-
-# def getDownload(v):
-# link = v.getDownloadURL('csv')
-# req = urllib.request.Request(url=link)
-# try:
-# handler = urllib.request.urlopen(req)
-# except HTTPError as e:
-# handler = e.read()
-# lines = []
-# for l in handler:
-# lines.append(str(l))
-# features_dem.append(lines)
-
-#--------------------------- DEM processing -------------------------------
-
-# Set collection
-dem_col = 'UMN/PGC/ArcticDEM/V3/2m_mosaic'
-
-# Get image collection
-scenes = getScene(dem_col, box)
-print(f'\nScenes gathered from {dem_col}')
-
-# Get elevation and slope
-elevation = getElevation(scenes)
-slope = getSlope(elevation)
-
-elevation = getSmooth(elevation, 110)
-
-# Mask out ocean pixels
-elevation = maskImage(elevation, ocean_mask)
-
-# Get sinks
-elevation = getInt(elevation)
-sinks = getSinks(elevation, 10, 50)
-
-# Remove speckle with smoothing
-sinks = getSmooth(sinks, 50).rename('dem_sinks')
-sinks = getInt(sinks)
-print(f'{dem_col} scenes classified')
-print('Band names: ' + str(sinks.bandNames().getInfo()))
-
-try:
- features_dem = getFeatures(sinks, 10, box)
-except:
- features_dem = getFeaturesSplit(sinks, 10, bbox)
-
-
-#--------------------------- SAR processing -------------------------------
-
-# Set collection
-sar_col = 'COPERNICUS/S1_GRD'
-
-# Get image collection
-scenes = getScenes(sar_col, date1, date2, box)
-if scenes.size().getInfo() > 0:
- scenes = filterSARscenes(scenes)
- print(f'\nScenes gathered from {sar_col}')
- print('Total number of scenes: ', scenes.size().getInfo())
- print('Number of bands per scene: '
- + str(len(scenes.first().bandNames().getInfo())))
- print('Band names: ' + str(scenes.first().bandNames().getInfo()))
-
- # Get average
- aver = getMosaic(scenes, 'HH')
-
- # Mask out ocean pixels
- aver = maskImage(aver, ocean_mask)
-
- # Classify water from SAR average image
- water_sar = classifySARimage(aver, -20, 'HH', 50)
- print(f'{sar_col} scenes classified')
- print('Band names: ' + str(water_sar.bandNames().getInfo()))
-
- try:
- features_sar = getFeatures(water_sar, 10, box)
- except:
- features_sar = getFeaturesSplit(water_sar, 10, bbox)
-
-else:
- features_sar = None
- print(f'\nNo {sar_col} scenes identified between {date1} and {date2}')
-
-#------------------------- VIS processing (S2) -----------------------------
-
-# Set collection
-vis_col1 = "COPERNICUS/S2"
-
-# Get image collection
-scenes = getScenes(vis_col1, date1, date2, box)
-if scenes.size().getInfo() > 0:
- scenes = filterS2scenes(scenes, cloud)
- print(f'\nScenes gathered from {vis_col1}')
- print('Total number of scenes: ', scenes.size().getInfo())
- print('Number of bands per scene: '
- + str(len(scenes.first().bandNames().getInfo())))
- print('Band names: ' + str(scenes.first().bandNames().getInfo()))
-
- # Mask scenes for clouds
- scenes = maskS2clouds(scenes)
- # ee.Terrain.hillshade(image, azimuth, elevation)
-
- # Get average of spectific bands
- scenes = renameS2scenes(scenes)
- scenes = resampleS2scenes(scenes)
- aver = getMean(scenes, ['blue','green','red','vnir','swir1_1','swir2_1'])
- print('Scenes resampled and mosiacked')
- print('Band names: ' + str(aver.bandNames().getInfo()))
-
- # Classify water from VIS average image, and mask out ocean pixels
- water_s2 = classifyVISimage(aver)
- water_s2 = maskImage(water_s2, ocean_mask)
- print(f'{vis_col1} scenes classified')
- print('Band names: ' + str(water_s2.bandNames().getInfo()))
-
- try:
- features_s2 = getFeatures(water_s2, 10, box)
- except:
- features_s2 = getFeaturesSplit(water_s2, 10, bbox)
-else:
- features_s2 = None
- print(f'\nNo {vis_col1} scenes identified between {date1} and {date2}')
-
-#----------------------- VIS processing (Landsat 8) -----------------------
-
-# Set collection
-vis_col2 = "LANDSAT/LC08/C01/T1_TOA"
-
-# Get image collection
-scenes = getScenes(vis_col2, date1, date2, box)
-if scenes.size().getInfo() > 0:
- scenes = filterLSscenes(scenes, cloud)
- print(f'\nScenes gathered from {vis_col2}')
- print('Total number of scenes: ', scenes.size().getInfo())
- print('Number of bands per scene: '
- + str(len(scenes.first().bandNames().getInfo())))
- print('Band names: ' + str(scenes.first().bandNames().getInfo()))
-
- # Mask scenes for clouds
- scenes = maskL8clouds(scenes)
- # ee.Terrain.hillshade(image, azimuth, elevation)
-
- # Get average of spectific bands
- scenes = renameL8scenes(scenes)
- aver = getMean(scenes, ['blue','green','red','vnir','swir1','swir2'])
- print('Scenes resampled and mosiacked')
- print('Band names: ' + str(aver.bandNames().getInfo()))
-
- # Classify water from VIS average image, and mask out ocean pixels
- ndwi = aver.normalizedDifference(['green_mean', 'vnir_mean']).rename('ndwi')
- mndwi = aver.normalizedDifference(['green_mean','swir1_mean']).rename('mndwi')
- aweish = aver.expression('BLUE + 2.5 * GREEN - 1.5 * (VNIR + SWIR1) - 0.25 * SWIR2',
- {'BLUE' : aver.select('blue_mean'),
- 'GREEN' : aver.select('green_mean'),
- 'SWIR1' : aver.select('swir1_mean'),
- 'VNIR' : aver.select('vnir_mean'),
- 'SWIR2' : aver.select('swir2_mean')}).rename('aweish')
- aweinsh = aver.expression('4.0 * (GREEN - SWIR1) - (0.25 * VNIR + 2.75 * SWIR2)',
- {'GREEN' : aver.select('green_mean'),
- 'SWIR1' : aver.select('swir1_mean'),
- 'VNIR' : aver.select('vnir_mean'),
- 'SWIR2' : aver.select('swir2_mean')}).rename('aweinsh')
- bright = aver.expression('(RED + GREEN + BLUE) / 3',
- {'BLUE' : aver.select('blue_mean'),
- 'GREEN' : aver.select('green_mean'),
- 'RED' : aver.select('red_mean')}).rename('bright')
-
- aver = aver.addBands([ndwi, mndwi, aweish, aweinsh, bright])
- classified = aver.expression("(BRIGHT > 5000) ? 0"
- ": (NDWI > 0.3) ? 1 "
- ": (MNDWI < 0.1) ? 0 "
- ": (AWEISH < 2000) ? 0"
- ": (AWEISH > 5000) ? 0"
- ": (AWEINSH < 4000) ? 0"
- ": (AWEINSH > 6000) ? 0"
- ": 1",
- {'NDWI' : aver.select('ndwi'),
- 'MNDWI' : aver.select('mndwi'),
- 'AWEISH' : aver.select('aweish'),
- 'AWEINSH' : aver.select('aweinsh'),
- 'BRIGHT' : aver.select('bright')}).rename('water')
- water_ls8 = classified.updateMask(classified)
- water_ls8 = maskImage(water_ls8, ocean_mask)
- print(f'{vis_col2} scenes classified')
- print('Band names: ' + str(water_ls8.bandNames().getInfo()))
-
- try:
- features_ls8 = getFeatures(water_ls8, 10, box)
- except:
- features_ls8 = getFeaturesSplit(water_ls8, 10, bbox)
-
-else:
- features_ls8 = None
- print(f'\nNo {vis_col2} scenes identified between {date1} and {date2}')
-
-
-#------------------------ Compile geodatabase -----------------------------
-
-iml = compileFeatures([features_s2, features_ls8, features_sar, features_dem],
- ['VIS','VIS','SAR','DEM'],
- [vis_col1, vis_col2, sar_col, dem_col],
- [date1, date2])
-print(f'\nCompiled {iml.shape[0]} geodatabase features')
-
-# Reproject geometries
-iml = iml.to_crs(proj)
-print(f'{iml.shape[0]} unfiltered features')
-
-iml.to_file(f"out/iiml_{date1}_{date2}_{aoi1[0]}_{aoi1[1]}_unfiltered.shp")
-
-
-#-------------------- Filter database by ice margin -----------------------
-
-# Load margin and add buffer
-margin = gpd.read_file("../../../other/datasets/ice_margin/gimp_icemask_line_polstereo.shp")
-margin_buff = margin.buffer(500)
-margin_buff = gpd.GeoDataFrame(geometry=margin_buff, crs=margin.crs)
-
-# Perform spatial join
-iml = gpd.sjoin(iml, margin_buff, how='left')
-iml = iml[iml['index_right']==0]
-iml = iml.drop(columns='index_right')
-print(f'{iml.shape[0]} features within 500 m of margin')
-
-# Filter lakes by area
-iml['area_sqkm'] = iml['geometry'].area/10**6
-iml['length_km'] = iml['geometry'].length/1000
-iml = iml[(iml.area_sqkm >= 0.05)]
-
-# Calculate geometry info
-iml.reset_index(inplace=True, drop=True)
-print(f'{iml.shape[0]} features over 0.05 sq km')
-iml.to_file(f"out/iiml_{date1}_{date2}_{aoi1[0]}_{aoi1[1]}_filtered.shp")
-
-#------------------------- Populate metadata ------------------------------
-
-iml = assignID(iml)
-iml = assignSources(iml)
-
-names = gpd.read_file('../../../other/datasets/placenames/oqaasileriffik_placenames.shp')
-iml = assignNames(iml, names)
-
-#----------------------- Write data to shapefile --------------------------
-
-iml.to_file(f"/home/pho/Desktop/python_workspace/GrIML/other/out/iiml_{date1}_{date2}_{aoi1[0]}_{aoi1[1]}_final.shp")
-print(f'Saved to out/iiml_{date1}_{date2}_{aoi1[0]}_{aoi1[1]}_final.shp')
-
-# Plot lakes
-iml.plot(figsize=(10, 10), alpha=0.5, edgecolor='k')
-
-print('\nFinished')
-end=time.time()
-print('Total run time: ' + strftime("%H:%M:%S", gmtime(round(end-start))))
diff --git a/src/griml/examples/getInventory_obj.py b/src/griml/examples/getInventory_obj.py
deleted file mode 100644
index f8b507c..0000000
--- a/src/griml/examples/getInventory_obj.py
+++ /dev/null
@@ -1,182 +0,0 @@
-"""
-GrIML processing module playground (experimental)
-
-@author: Penelope How
-"""
-
-import time,sys
-from time import gmtime, strftime
-import geopandas as gpd
-
-try:
- sys.path.append('../')
- from process import gee
- from lake import dissolvePolygons, compileFeatures, assignID, assignSources, assignNames
-except:
- from griml.process import gee
- from griml.lake import dissolvePolygons, compileFeatures, assignID, \
- assignSources, assignNames
-
-start=time.time()
-
-#------------------------------------------------------------------------------
-
-# Set AOI box splitter parameters
-wh=0.3 # Window height
-ww=0.3 # Window width
-oh = 0.05 # Overlap height
-ow = 0.05 # Overlap width
-
-# Set date range
-date1='2017-07-01'
-date2='2017-08-31'
-
-# Set output projection
-proj = 'EPSG:3413' # Polar stereographic
-
-# Set AOI
-# aoi = '/home/pho/python_workspace/GrIML/other/datasets/aoi/AOI_mask_split2.shp'
-aoi = '/home/pho/python_workspace/GrIML/other/datasets/aoi/test_mask_4326.shp'
-aoi = gpd.read_file(aoi)#.to_crs('EPSG:4326')
-
-# Load ice margin with buffer
-print('Preparing ice margin buffer...')
-margin_buff = gpd.read_file("/home/pho/python_workspace/GrIML/other/datasets/ice_margin/gimp_icemask_line_polstereo_simple_buffer.shp")
-# margin_buff = margin.buffer(500)
-# margin_buff = gpd.GeoDataFrame(geometry=margin_buff, crs=margin.crs)
-
-# Load name database
-print('Loading placename database...')
-names = gpd.read_file('/home/pho/python_workspace/GrIML/other/datasets/placenames/oqaasileriffik_placenames.shp')
-
-
-#--------------------------- Initialise GEE -------------------------------
-
-parameters = [#{'collection' : 'UMN/PGC/ArcticDEM/V3/2m_mosaic',
- #'smooth' : 100, 'fill' : 100, 'kernel' : 100, 'speckle' : 50},
- # {'collection' : 'UMN/PGC/ArcticDEM/V3/2m',
- # 'smooth' : 100, 'fill' : 100, 'kernel' : 100, 'speckle' : 50},
- {'collection' : 'COPERNICUS/S1_GRD',
- 'polar' : 'HH', 'threshold' : -20, 'smooth' : 50},
- #{'collection' : 'COPERNICUS/S2', `
- #'cloud' : 20},
- {'collection' : 'LANDSAT/LC08/C01/T1_TOA',
- 'cloud' : 20}]
- # {'collection' : 'LANDSAT/LE07/C02/T1_TOA',
- # 'cloud' : 50}]
-
-
-
-for i in range(len(aoi.index)):
- print(f'\nCommencing processing of {list(aoi["catch"])[i]} catchment area')
- aoi_poly = aoi.geometry.iloc[i]
- xmin, ymin, xmax, ymax = aoi_poly.bounds
-
- print('Clipping margin to catchment area...')
- aoi_poly_proj = gpd.GeoDataFrame(geometry=[aoi_poly], crs=aoi.crs).to_crs(proj)
- margin_clip = gpd.clip(margin_buff, aoi_poly_proj)
-
- print('Conducting classifications...')
- proc = gee([date1, date2], aoi_poly, parameters, [wh, ww, oh, ow], True)
-
- print('Processing...')
- water = proc.processAll()
-
- print('Retrieving...')
- features = proc.retrieveAll(water)
-
- # Filter features
- filtered=[]
- for f in features:
- print('Filtering features...')
-
- # Remove duplicates
- print('Dissolving polygons...')
- gdf = gpd.GeoDataFrame(geometry=f, crs='EPSG:4326')
- gdf = dissolvePolygons(gdf)
-
- # Remove lakes below size threshold
- print('Filtering by size...')
- gdf = gdf.set_crs('EPSG:4326')
- gdf = gdf.to_crs(proj)
- gdf['area_sqkm'] = gdf['geometry'].area/10**6
- gdf = gdf[(gdf.area_sqkm >= 0.05)]
-
- # # Remove lakes outside of margin boundary
- # print('Filtering by bounds...')
- # gdf = gpd.sjoin(gdf, margin_clip, how='left')
- # gdf = gdf[gdf['index_right']==0]
- # gdf = gdf.drop(columns='index_right')
- filtered.append(gdf.geometry)
-
-
-#------------------------ Compile geodatabase -----------------------------
-# lakes=[]
-# for f in features:
-# gdf = gpd.GeoDataFrame(geometry=f, crs='EPSG:4326')
-# gdf = dissolvePolygons(gdf)
-# gdf['area'] = gdf['geometry'].area/10**6
-# gdf = gdf[(gdf.area >= 0.05)]
-
-# gdf = gpd.sjoin(gdf, margin_buff, how='left')
-# gdf = gdf[gdf['index_right']==0]
-# gdf = gdf.drop(columns='index_right')
-# lakes.append(list(gdf['geometry']))
-
- # Compile all features
- iml = compileFeatures(filtered,
- ['DEM', 'SAR', 'VIS'],
- ['UMN/PGC/ArcticDEM/V3/2m_mosaic', 'COPERNICUS/S1_GRD',
- 'LANDSAT/LC08/C01/T1_TOA'],
- [date1, date2], proj)
- print(f'\nCompiled {iml.shape[0]} geodatabase features')
-
- # Calculate geometry attributes
- iml['area_sqkm'] = iml['geometry'].area/10**6
- iml['length_km'] = iml['geometry'].length/1000
- iml = iml[(iml.area_sqkm >= 0.05)]
- iml['catch'] = str(list(aoi["catch"])[i])
-
- # Calculate geometry info
- iml.reset_index(inplace=True, drop=True)
-
- print(f'{iml.shape[0]} filtered features')
-
- iml.to_file(f'out/iiml_{date1}_{date2}_{list(aoi["catch"])[i]}_filtered.shp')
-
-
-# #-------------------- Filter database by ice margin -----------------------
-
-# # Perform spatial join
-# aoi_poly_proj = aoi_poly.to_crs(proj)
-# margin_clip = gpd.clip(margin_buff, aoi_poly_proj)
-
-# iml = gpd.sjoin(iml, margin_clip, how='left')
-# iml = iml[iml['index_right']==0]
-# iml = iml.drop(columns='index_right')
-# print(f'{iml.shape[0]} features within 500 m of margin')
-
-# # Calculate geometry info
-# iml.reset_index(inplace=True, drop=True)
-# print(f'{iml.shape[0]} features over 0.05 sq km')
-# iml.to_file(f'out/iiml_{date1}_{date2}_{list(aoi["catch"])[i]}_filtered.shp')
-
-
-#-------------------- Populate metadata and write to file -----------------
-
-# iml = assignID(iml)
- # iml = assignSources(iml)
-
- # iml = assignNames(iml, names)
-
- # iml.to_file(f'out/iiml_{date1}_{date2}_{list(aoi["catch"])[i]}_final.shp')
- # print(f'Saved to out/iiml_{date1}_{date2}_{list(aoi["catch"])[i]}_final.shp')
-
- # # Plot lakes
- # iml.plot(figsize=(10, 10), alpha=0.5, edgecolor='k')
-
-
-#------------------------------------------------------------------------------
-print('\nFinished')
-end=time.time()
-print('Total run time: ' + strftime("%H:%M:%S", gmtime(round(end-start))))
diff --git a/src/griml/metadata/__init__.py b/src/griml/metadata/__init__.py
index 3d16982..7e1e3d8 100644
--- a/src/griml/metadata/__init__.py
+++ b/src/griml/metadata/__init__.py
@@ -1,5 +1,6 @@
from griml.metadata.assign_certainty import *
from griml.metadata.assign_id import *
from griml.metadata.assign_names import *
+from griml.metadata.assign_regions import *
from griml.metadata.assign_sources import *
from griml.metadata.add_metadata import *
diff --git a/src/griml/metadata/add_metadata.py b/src/griml/metadata/add_metadata.py
index 2f15ee2..50e6d9a 100644
--- a/src/griml/metadata/add_metadata.py
+++ b/src/griml/metadata/add_metadata.py
@@ -1,30 +1,17 @@
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
-from griml.metadata import assign_id, assign_sources, assign_certainty, \
- assign_names
from griml.load import load
+from griml.metadata import assign_id, assign_sources, assign_certainty, \
+ assign_names, assign_regions
import geopandas as gpd
-def add_metadata(iml_file, names_file, outfile=None):
- '''Add metadata to collection of features
-
- Parameters
- ----------
- iml_file : str, geopandas.dataframe.DataFrame
- Filepath or geopandas.dataframe.DataFrame object to assign metadata to
-
- Returns
- -------
- iml: geopandas.dataframe.GeoDataFrame
- Geodataframe with metadata
- '''
-
- # Load files
- iml = load(iml_file)
- names = load(names_file)
+def add_metadata(iml, names, regions, outfile=None):
+
+ iml = load(iml)
+ names = load(names)
+ regions = load(regions)
- # Perform metadata steps
print('Assigning ID...')
iml = assign_id(iml)
@@ -35,24 +22,27 @@ def add_metadata(iml_file, names_file, outfile=None):
n = ['S1','S2','ARCTICDEM']
scores = [0.298, 0.398, 0.304]
iml = assign_certainty(iml, n, scores)
+
+ print('Assigning regions...')
+ iml = assign_regions(iml, regions)
print('Assigning placenames...')
iml = assign_names(iml, names)
- # Save to file if given
- if outfile is not None:
+ if outfile:
print('Saving file...')
iml.to_file(outfile)
- print('Saved to '+str(outfile))
-
- return iml
+ print('Saved to '+str(outfile)+"_metadata.shp")
if __name__ == "__main__":
- infile1 = '../test/test_merge_2.shp'
- infile2 = '../test/test_placenames.shp'
- add_metadata(infile1, infile2)
+ # indir = "/home/pho/python_workspace/GrIML/other/iml_2017/merged_vectors/griml_2017_inventory.shp"
+ indir = "/home/pho/python_workspace/GrIML/other/iml_2017/inspected_vectors/lakes_all-0000000000-0000037888_filtered.shp"
+
+
+ infile_names = '/home/pho/python_workspace/GrIML/other/datasets/placenames/oqaasileriffik_placenames.shp'
+ infile_regions = '/home/pho/python_workspace/GrIML/other/datasets/drainage_basins/greenland_basins_polarstereo.shp'
+
- iml = gpd.read_file(infile1)
- names = gpd.read_file(infile2)
- add_metadata(iml, names)
\ No newline at end of file
+ outfile = "/home/pho/python_workspace/GrIML/other/iml_2017/metadata_vectors/griml_2017_inventory_final.shp"
+ add_metadata(indir, infile_names, infile_regions, outfile)
diff --git a/src/griml/metadata/assign_names.py b/src/griml/metadata/assign_names.py
index 9bd294f..c151a48 100644
--- a/src/griml/metadata/assign_names.py
+++ b/src/griml/metadata/assign_names.py
@@ -1,7 +1,18 @@
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
-def assign_names(gdf, gdf_names, distance=500.0):
+import itertools
+from operator import itemgetter
+
+import geopandas as gpd
+import numpy as np
+import pandas as pd
+
+from scipy.spatial import cKDTree
+from shapely.geometry import Point, LineString, Polygon
+from griml.load import load
+
+def assign_names(gdf, gdf_names):
'''Assign placenames to geodataframe geometries based on names in another
geodataframe point geometries
@@ -19,62 +30,41 @@ def assign_names(gdf, gdf_names, distance=500.0):
gdf : pandas.GeoDataFrame
Vectors with assigned IDs
'''
- placenames = _compile_names(gdf_names)
+
+ # Load geodataframes
+ gdf1 = load(gdf)
+ gdf2 = load(gdf_names)
+
+ # Compile placenames into new dataframe
+ names = _compile_names(gdf2)
+ placenames = gpd.GeoDataFrame({"geometry": list(gdf2['geometry']),
+ "placename": names})
- lakename=[]
- for i,v in gdf.iterrows():
-
- geom = v['geometry']
-
- polynames=[]
- for pt in range(len(placenames)):
- if geom.contains(gdf_names['geometry'][pt]) == True:
- polynames.append(placenames[pt])
-
- if len(polynames)==0:
- for pt in range(len(placenames)):
- if gdf_names['geometry'][pt].distance(geom) < distance:
- polynames.append(placenames[pt])
- lakename.append(polynames)
-
- elif len(polynames)==1:
- lakename.append(polynames)
-
- else:
- out=[]
- for p in polynames:
- out.append(p)
- lakename.append(out)
-
- lakeid = gdf['unique_id'].tolist()
- lakename2 = []
+ # Assign names based on proximity
+ a = _get_nearest_point(gdf1, placenames)
+ return a
+
+
+def _get_nearest_point(gdA, gdB, distance=500.0):
+ '''Return properties of nearest point in Y to geometry in X'''
+ nA = np.array(list(gdA.geometry.centroid.apply(lambda x: (x.x, x.y))))
+ nB = np.array(list(gdB.geometry.apply(lambda x: (x.x, x.y))))
+
+ btree = cKDTree(nB)
+ dist, idx = btree.query(nA, k=1)
+ gdB_nearest = gdB.iloc[idx].drop(columns="geometry").reset_index(drop=True)
+ gdf = pd.concat(
+ [
+ gdA.reset_index(drop=True),
+ gdB_nearest,
+ pd.Series(dist, name='dist')
+ ],
+ axis=1)
- for x in gdf.index:
- indx = _get_indices(lakeid, x)
- findname=[]
- for l in indx:
- if len(lakename[l])!=0:
- findname.append(lakename[l])
-
- for i in range(len(indx)):
- if len(findname)==0:
- lakename2.append('')
-
- else:
- unique = set(findname[0])
- unique = list(unique)
-
- if len(unique)==1:
- lakename2.append(findname[0][0])
-
- else:
- out2 = ', '
- out2 = out2.join(unique)
- lakename2.append(out2)
- gdf['placename'] = lakename2
+ gdf.loc[gdf['dist']>=distance, 'placename'] = 'Unknown'
+ gdf = gdf.drop(columns=['dist'])
return gdf
-
def _get_indices(mylist, value):
'''Get indices for value in list'''
return[i for i, x in enumerate(mylist) if x==value]
@@ -95,3 +85,14 @@ def _compile_names(gdf):
else:
placenames.append(None)
return placenames
+
+
+if __name__ == "__main__":
+
+ # Define file attributes
+ infile = '/home/pho/Desktop/python_workspace/GrIML/src/griml/test/test_merge_1.shp'
+ names = '/home/pho/Desktop/python_workspace/GrIML/other/datasets/placenames/oqaasileriffik_placenames_polarstereo.shp'
+
+
+ # Perform conversion
+ a = assign_names(infile, names)
\ No newline at end of file
diff --git a/src/griml/metadata/assign_region.py b/src/griml/metadata/assign_region.py
deleted file mode 100644
index 9bd294f..0000000
--- a/src/griml/metadata/assign_region.py
+++ /dev/null
@@ -1,97 +0,0 @@
-#!/usr/bin/env python3
-# -*- coding: utf-8 -*-
-
-def assign_names(gdf, gdf_names, distance=500.0):
- '''Assign placenames to geodataframe geometries based on names in another
- geodataframe point geometries
-
- Parameters
- ----------
- gdf : pandas.GeoDataFrame
- Vectors to assign uncertainty to
- gdf_names : pandas.GeoDataFrame
- Vector geodataframe with placenames
- distance : int
- Distance threshold between a given vector and a placename
-
- Returns
- -------
- gdf : pandas.GeoDataFrame
- Vectors with assigned IDs
- '''
- placenames = _compile_names(gdf_names)
-
- lakename=[]
- for i,v in gdf.iterrows():
-
- geom = v['geometry']
-
- polynames=[]
- for pt in range(len(placenames)):
- if geom.contains(gdf_names['geometry'][pt]) == True:
- polynames.append(placenames[pt])
-
- if len(polynames)==0:
- for pt in range(len(placenames)):
- if gdf_names['geometry'][pt].distance(geom) < distance:
- polynames.append(placenames[pt])
- lakename.append(polynames)
-
- elif len(polynames)==1:
- lakename.append(polynames)
-
- else:
- out=[]
- for p in polynames:
- out.append(p)
- lakename.append(out)
-
- lakeid = gdf['unique_id'].tolist()
- lakename2 = []
-
- for x in gdf.index:
- indx = _get_indices(lakeid, x)
- findname=[]
- for l in indx:
- if len(lakename[l])!=0:
- findname.append(lakename[l])
-
- for i in range(len(indx)):
- if len(findname)==0:
- lakename2.append('')
-
- else:
- unique = set(findname[0])
- unique = list(unique)
-
- if len(unique)==1:
- lakename2.append(findname[0][0])
-
- else:
- out2 = ', '
- out2 = out2.join(unique)
- lakename2.append(out2)
- gdf['placename'] = lakename2
- return gdf
-
-
-def _get_indices(mylist, value):
- '''Get indices for value in list'''
- return[i for i, x in enumerate(mylist) if x==value]
-
-
-def _compile_names(gdf):
- '''Get preferred placenames from placename geodatabase'''
- placenames=[]
- for i,v in gdf.iterrows():
- if v['Ny_grønla'] != None:
- placenames.append(v['Ny_grønla'])
- else:
- if v['Dansk'] != None:
- placenames.append(v['Dansk'])
- else:
- if v['Alternativ'] != None:
- placenames.append(v['Alternativ'])
- else:
- placenames.append(None)
- return placenames
diff --git a/src/griml/metadata/assign_regions.py b/src/griml/metadata/assign_regions.py
new file mode 100644
index 0000000..6458989
--- /dev/null
+++ b/src/griml/metadata/assign_regions.py
@@ -0,0 +1,86 @@
+#!/usr/bin/env python3
+# -*- coding: utf-8 -*-
+
+import itertools
+from operator import itemgetter
+
+import geopandas as gpd
+import numpy as np
+import pandas as pd
+
+from griml.load import load
+from scipy.spatial import cKDTree
+from shapely.geometry import Point, LineString, Polygon
+
+def assign_regions(gdf, gdf_regions, region_name='region'):
+ '''Assign region to geodataframe geometries based on regions in another
+ geodataframe object
+
+ Parameters
+ ----------
+ gdf : pandas.GeoDataFrame
+ Vectors to assign region name to
+ gdf_regions : pandas.GeoDataFrame
+ Vector geodataframe with regions
+
+ Returns
+ -------
+ gdf : pandas.GeoDataFrame
+ Vectors with assigned IDs
+ '''
+
+ # Load geodataframes
+ gdf1 = load(gdf)
+ gdf2 = load(gdf_regions)
+
+ g = _get_nearest_polygon(gdf1, gdf2)
+
+ return g
+
+
+def _get_nearest_polygon(gdfA, gdfB, gdfB_cols=['subregion'], distance=100000.0):
+ '''Return given properties of nearest polygon in Y to geometry in X'''
+
+ A = np.array(list(gdfA.geometry.centroid.apply(lambda x: (x.x, x.y))))
+ # B = [np.array(geom.boundary.coords) for geom in gdfB.geometry.to_list()]
+
+ B=[]
+ for geom in gdfB.geometry.to_list():
+ try:
+ g = np.array(geom.boundary.coords)
+ except:
+ g = np.array(geom.boundary.geoms[0].coords)
+ B.append(g)
+
+
+ B_ix = tuple(itertools.chain.from_iterable(
+ [itertools.repeat(i, x) for i, x in enumerate(list(map(len, B)))]))
+ B = np.concatenate(B)
+ ckd_tree = cKDTree(B)
+
+ dist, idx = ckd_tree.query(A, k=1)
+ idx = itemgetter(*idx)(B_ix)
+
+ gdf = pd.concat(
+ [gdfA, gdfB.loc[idx, gdfB_cols].reset_index(drop=True),
+ pd.Series(dist, name='dist')], axis=1)
+
+ gdf.loc[gdf['dist']>=distance, 'subregion'] = 'Unknown'
+
+ gdf = gdf.drop(columns=['dist'])
+ return gdf
+
+
+if __name__ == "__main__":
+
+ # Define file attributes
+ infile = '/home/pho/Desktop/python_workspace/GrIML/src/griml/test/test_merge_1.shp'
+ regions = '/home/pho/python_workspace/GrIML/other/datasets/drainage_basins/greenland_basins_polarstereo.shp'
+ regions=gpd.read_file(regions)
+
+ regions_dissolve = regions.dissolve(by='subregion')
+
+
+ # Perform conversion
+ a = assign_regions(infile, regions)
+
diff --git a/src/griml/metadata/metadata.py b/src/griml/metadata/metadata.py
deleted file mode 100644
index 9b0ca30..0000000
--- a/src/griml/metadata/metadata.py
+++ /dev/null
@@ -1,38 +0,0 @@
-#!/usr/bin/env python3
-# -*- coding: utf-8 -*-
-
-from griml.metadata import assign_id, assign_sources, assign_certainty, \
- assign_names
-import geopandas as gpd
-
-def add_metadata(iml, names, outfile):
-
- print('Assigning ID...')
- iml = assign_id(iml)
-
- print('Assigning sources...')
- iml = assign_sources(iml)
-
- print('Assigning certainty scores...')
- n = ['S1','S2','ARCTICDEM']
- scores = [0.298, 0.398, 0.304]
- iml = assign_certainty(iml, n, scores)
-
- print('Assigning placenames...')
- iml = assign_names(iml, names)
-
- print('Saving file...')
- iml.to_file(outfile)
- print('Saved to '+str(outfile)+"_metadata.shp")
-
-
-if __name__ == "__main__":
- # indir = "/home/pho/python_workspace/GrIML/other/iml_2017/merged_vectors/griml_2017_inventory.shp"
- indir = "/home/pho/python_workspace/GrIML/other/iml_2017/inspected_vectors/lakes_all-0000000000-0000037888_filtered.shp"
- iml = gpd.read_file(indir)
-
- infile_names = '/home/pho/python_workspace/GrIML/other/datasets/placenames/oqaasileriffik_placenames.shp'
- names = gpd.read_file(infile_names)
-
- outfile = "/home/pho/python_workspace/GrIML/other/iml_2017/metadata_vectors/griml_2017_inventory_final.shp"
- add_metadata(iml, names, outfile)
diff --git a/src/griml/test/greenland_basins_polarstereo.cpg b/src/griml/test/greenland_basins_polarstereo.cpg
new file mode 100644
index 0000000..cd89cb9
--- /dev/null
+++ b/src/griml/test/greenland_basins_polarstereo.cpg
@@ -0,0 +1 @@
+ISO-8859-1
\ No newline at end of file
diff --git a/src/griml/test/greenland_basins_polarstereo.dbf b/src/griml/test/greenland_basins_polarstereo.dbf
new file mode 100644
index 0000000..9d97cd3
Binary files /dev/null and b/src/griml/test/greenland_basins_polarstereo.dbf differ
diff --git a/src/griml/test/greenland_basins_polarstereo.prj b/src/griml/test/greenland_basins_polarstereo.prj
new file mode 100644
index 0000000..2e49174
--- /dev/null
+++ b/src/griml/test/greenland_basins_polarstereo.prj
@@ -0,0 +1 @@
+PROJCS["WGS_1984_NSIDC_Sea_Ice_Polar_Stereographic_North",GEOGCS["GCS_WGS_1984",DATUM["D_WGS_1984",SPHEROID["WGS_1984",6378137.0,298.257223563]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Stereographic_North_Pole"],PARAMETER["False_Easting",0.0],PARAMETER["False_Northing",0.0],PARAMETER["Central_Meridian",-45.0],PARAMETER["Standard_Parallel_1",70.0],UNIT["Meter",1.0]]
\ No newline at end of file
diff --git a/src/griml/test/greenland_basins_polarstereo.shp b/src/griml/test/greenland_basins_polarstereo.shp
new file mode 100644
index 0000000..2773592
Binary files /dev/null and b/src/griml/test/greenland_basins_polarstereo.shp differ
diff --git a/src/griml/test/greenland_basins_polarstereo.shx b/src/griml/test/greenland_basins_polarstereo.shx
new file mode 100644
index 0000000..a63669b
Binary files /dev/null and b/src/griml/test/greenland_basins_polarstereo.shx differ
diff --git a/src/griml/test/process_cli.py b/src/griml/test/process_cli.py
new file mode 100644
index 0000000..915effc
--- /dev/null
+++ b/src/griml/test/process_cli.py
@@ -0,0 +1,132 @@
+#!/usr/bin/env python3
+# -*- coding: utf-8 -*-
+"""
+@author: pho
+"""
+from griml.convert import convert
+from griml.filter import filter_vectors
+from griml.merge import merge_vectors
+from griml.metadata import add_metadata
+from pathlib import Path
+import glob, os
+from argparse import ArgumentParser
+
+def parse_griml_arguments():
+ parser = ArgumentParser(description="Full post-processing workflow for "+
+ "creating ice marginal lake inventory")
+ parser.add_argument('-r', '--root_dir', type=str, required=True,
+ help='Root directory to write files to')
+ parser.add_argument('-i', '--in_dir', type=str, required=True,
+ help='Directory path to input raster files')
+ parser.add_argument('-y', '--year', type=str, required=True,
+ help='Year of inventory')
+ parser.add_argument('-m', '--margin_file', type=str, required=True,
+ help='File path to ice margin for spatial filtering')
+ parser.add_argument('-n', '--names_file', type=str, required=True,
+ help='File path to placenames file for metadata population')
+ parser.add_argument('-p', '--proj', type=str, default='EPSG:3413',
+ required=False, help='Projection (of input and output)')
+ parser.add_argument('-s', '--steps', type=str, default='1111',
+ required=False, help='Define which steps to include in'+
+ ' processing, where each value indicates: convert, '+
+ 'filter, merge, and metadata. If set to zero, the '+
+ 'step associated with that position is skipped')
+
+ args = parser.parse_args()
+ return args
+
+def get_step_flags(a):
+ '''Return step flags'''
+ return a[0],a[1],a[2],a[3]
+
+
+def check_dir(d):
+ '''Check if directory exists and create it if it does not'''
+ if not os.path.exists(d):
+ os.mkdir(d)
+
+
+def griml_process():
+ '''Perform processing workflow'''
+ args = parse_griml_arguments()
+
+ s1, s2, s3, s4 = get_step_flags(args.steps)
+
+ print('Commencing post processing for inventory year ' + args.year)
+ print('Adopted projection: ' + args.proj)
+ print('Writing outputs to ' + args.root_dir)
+
+ root_dir = Path(args.root_dir)
+
+ # Convert to vectors
+ if s1:
+ print('Converting rasters to vectors...')
+
+ src=args.in_dir
+ dest = str(root_dir.joinpath('vectors'))
+ check_dir(dest)
+
+ print('Reading from ' + src)
+ print('Writing to ' + dest)
+
+ band_info = [{'b_number':1, 'method':'VIS', 'source':'S2'},
+ {'b_number':2, 'method':'SAR', 'source':'S1'},
+ {'b_number':3, 'method':'DEM', 'source':'ARCTICDEM'}]
+ start=args.year+'0701'
+ end=args.year+'0831'
+
+ infiles = list(glob.glob(src+'/*.tif'))
+
+ convert(infiles, args.proj, band_info, start, end, str(dest))
+
+
+ # Filter vectors by area and proximity to margin
+ if s2:
+ print('Filtering vectors...')
+
+ src = str(root_dir.joinpath('vectors'))
+ dest = str(root_dir.joinpath('filtered'))
+ check_dir(dest)
+
+ print('Reading from ' + src)
+ print('Writing to ' + dest)
+
+ # margin_buff = gpd.read_file(infile_margin)
+ # margin_buff = margin.buffer(500)
+ # margin_buff = gpd.GeoDataFrame(geometry=margin_buff, crs=margin.crs)
+
+ infiles = list(glob.glob(src+'/*.shp'))
+
+ filter_vectors(infiles, args.margin_file, dest)
+
+
+ # Merge vectors
+ if s3:
+ print('Merging vectors...')
+
+ src = str(root_dir.joinpath('filtered'))
+ dest = root_dir.joinpath('merged/'+args.year+'_merged.shp')
+ check_dir(dest.parent)
+
+ print('Reading from ' + src)
+ print('Writing to ' + str(dest))
+
+ infiles = list(glob.glob(src+'/*.shp'))
+
+ merge_vectors(infiles, str(dest))
+
+
+ # Add metadata
+ if s4:
+ print('Adding metadata...')
+
+ src = str(root_dir.joinpath('merged/'+args.year+'_merged.shp'))
+ dest = root_dir.joinpath('metadata/'+args.year+'_metadata.shp')
+ check_dir(dest.parent)
+
+ print('Reading from ' + src)
+ print('Writing to ' + str(dest))
+
+ add_metadata(src, args.names_file, str(dest))
+
+ print('Finished')
diff --git a/src/griml/test/process_example.py b/src/griml/test/process_example.py
new file mode 100644
index 0000000..c96a4d9
--- /dev/null
+++ b/src/griml/test/process_example.py
@@ -0,0 +1,85 @@
+#!/usr/bin/env python3
+# -*- coding: utf-8 -*-
+"""
+@author: pho
+"""
+from griml.convert import convert
+from griml.filter import filter_vectors
+from griml.merge import merge_vectors
+from griml.metadata import add_metadata
+from pathlib import Path
+import glob
+
+root_dir = Path('/home/pho/python_workspace/GrIML/other/')
+year='2016'
+
+print('Commencing post processing for inventory year ' + year)
+
+# Convert to vectors
+print('Converting rasters to vectors...')
+
+src = str(root_dir.joinpath('iml_2016-2023/rasters/'+year+'_iml'))
+dest = str(root_dir.joinpath('iml_2016-2023/vectors/'+year))
+
+print('Reading from ' + src)
+print('Writing to ' + dest)
+
+proj = 'EPSG:3413'
+band_info = [{'b_number':1, 'method':'VIS', 'source':'S2'},
+ {'b_number':2, 'method':'SAR', 'source':'S1'},
+ {'b_number':3, 'method':'DEM', 'source':'ARCTICDEM'}]
+start=year+'0701'
+end=year+'0831'
+
+infiles = list(glob.glob(src+'/*.tif'))
+
+convert(infiles, proj, band_info, start, end, str(dest))
+
+
+# Filter vectors by area and proximity to margin
+print('Filtering vectors...')
+
+src = dest
+dest = str(root_dir.joinpath('iml_2016-2023/filtered/'+year))
+infile_margin = str(root_dir.joinpath('datasets/ice_margin/gimp_icemask_line_polstereo_simple_buffer.shp'))
+
+print('Reading from ' + src)
+print('Writing to ' + dest)
+
+# margin_buff = gpd.read_file(infile_margin)
+# margin_buff = margin.buffer(500)
+# margin_buff = gpd.GeoDataFrame(geometry=margin_buff, crs=margin.crs)
+
+infiles = list(glob.glob(src+'/*.shp'))
+
+filter_vectors(infiles, infile_margin, dest)
+
+
+# Merge vectors
+print('Merging vectors...')
+
+src = dest
+dest = str(root_dir.joinpath('iml_2016-2023/merged/'+year+'_merged.shp'))
+
+print('Reading from ' + src)
+print('Writing to ' + dest)
+
+infiles = list(glob.glob(src+'/*.shp'))
+
+merge_vectors(infiles, dest)
+
+
+# Add metadata
+print('Adding metadata...')
+
+src = dest
+dest = str(root_dir.joinpath('iml_2016-2023/metadata/'+year+'_metadata.shp'))
+
+print('Reading from ' + src)
+print('Writing to ' + dest)
+
+infile_names = str(root_dir.joinpath('datasets/placenames/oqaasileriffik_placenames.shp'))
+
+add_metadata(src, infile_names, dest)
+
+print('Finished')
diff --git a/src/griml/test/test.py b/src/griml/test/test.py
index 336fdee..24346c3 100644
--- a/src/griml/test/test.py
+++ b/src/griml/test/test.py
@@ -17,7 +17,6 @@ def test_convert(self):
{'b_number':3, 'method':'DEM', 'source':'ARCTICDEM'}]
start='20170701'
end='20170831'
-
infile = os.path.join(os.path.dirname(griml.__file__),'test/test_north_greenland.tif')
convert([infile], proj, band_info, start, end)
@@ -37,7 +36,8 @@ def test_metadata(self):
'''Test metadata population'''
infile1 = os.path.join(os.path.dirname(griml.__file__),'test/test_merge_2.shp')
infile2 = os.path.join(os.path.dirname(griml.__file__),'test/test_placenames.shp')
- add_metadata(infile1, infile2)
+ infile3 = os.path.join(os.path.dirname(griml.__file__),'test/greenland_basins_polarstereo.shp')
+ add_metadata(infile1, infile2, infile3)
if __name__ == "__main__":
unittest.main()
 +
+ **GrIML** proposes to examine ice marginal lake changes across Greenland using a multi-sensor and multi-method remote sensing approach to better address their influence on sea level contribution forecasting.
-Ice marginal lakes are detected using a remote sensing approach, based on offline workflows developed within the [ESA Glaciers CCI](https://catalogue.ceda.ac.uk/uuid/7ea7540135f441369716ef867d217519") (Option 6, An Inventory of Ice-Marginal Lakes in Greenland) ([How et al., 2021](https://www.nature.com/articles/s41598-021-83509-1)). Lake extents are defined through a multi-sensor approach using:
+Ice marginal lakes are detected using a remote sensing approach, based on offline workflows developed within the [ESA Glaciers CCI](https://catalogue.ceda.ac.uk/uuid/7ea7540135f441369716ef867d217519") (Option 6, An Inventory of Ice-Marginal Lakes in Greenland) ([How et al., 2021](https://www.nature.com/articles/s41598-021-83509-1)). Initial classifications are performed on Google Earth Engine with the scripts available [here](https://github.com/PennyHow/GrIML/tree/main/gee_scripts). Lake extents are defined through a multi-sensor approach using:
- Multi-spectral indices classification from Sentinel-2 optical imagery
- Backscatter classification from Sentinel-1 SAR (synthetic aperture radar) imagery
- Sink detection from ArcticDEM digital elevation models
+Post-processing of these classifications is performed using the GrIML post-processing Python package, including raster-to-vector conversion, filtering, merging, metadata population, and statistical analysis.
## Project links
diff --git a/src/griml.egg-info/PKG-INFO b/src/griml.egg-info/PKG-INFO
deleted file mode 100644
index f83ffdc..0000000
--- a/src/griml.egg-info/PKG-INFO
+++ /dev/null
@@ -1,76 +0,0 @@
-Metadata-Version: 2.1
-Name: griml
-Version: 0.0.1
-Summary: A workflow for classifying lakes from satellite imagery and compiling lake inventories
-Home-page: https://github.com/PennyHow/GrIML
-Author: Penelope How
-Author-email: pho@geus.dk
-License: UNKNOWN
-Project-URL: Bug Tracker, https://github.com/PennyHow/GrIML/issues
-Keywords: glaciology ice lake ESA
-Platform: UNKNOWN
-Classifier: Programming Language :: Python :: 3
-Classifier: License :: OSI Approved :: MIT License
-Classifier: Development Status :: 4 - Beta
-Classifier: Intended Audience :: Science/Research
-Classifier: Natural Language :: English
-Classifier: Topic :: Scientific/Engineering
-Classifier: Operating System :: OS Independent
-Requires-Python: >=3.6
-Description-Content-Type: text/markdown
-License-File: LICENSE
-
-# Investigating Greenland's ice marginal lakes under a changing climate (GrIML)
-
-[](https://griml.readthedocs.io/en/latest/?badge=latest)
-
-A repository for all project-related materials, funded under the ESA Living Planet Fellowship.
-
-**Project aim:** To examine ice marginal lake changes across Greenland using a multi-method and multi-sensor remote sensing approach, refined with in situ validation.
-
-## Background
-
-
**GrIML** proposes to examine ice marginal lake changes across Greenland using a multi-sensor and multi-method remote sensing approach to better address their influence on sea level contribution forecasting.
-Ice marginal lakes are detected using a remote sensing approach, based on offline workflows developed within the [ESA Glaciers CCI](https://catalogue.ceda.ac.uk/uuid/7ea7540135f441369716ef867d217519") (Option 6, An Inventory of Ice-Marginal Lakes in Greenland) ([How et al., 2021](https://www.nature.com/articles/s41598-021-83509-1)). Lake extents are defined through a multi-sensor approach using:
+Ice marginal lakes are detected using a remote sensing approach, based on offline workflows developed within the [ESA Glaciers CCI](https://catalogue.ceda.ac.uk/uuid/7ea7540135f441369716ef867d217519") (Option 6, An Inventory of Ice-Marginal Lakes in Greenland) ([How et al., 2021](https://www.nature.com/articles/s41598-021-83509-1)). Initial classifications are performed on Google Earth Engine with the scripts available [here](https://github.com/PennyHow/GrIML/tree/main/gee_scripts). Lake extents are defined through a multi-sensor approach using:
- Multi-spectral indices classification from Sentinel-2 optical imagery
- Backscatter classification from Sentinel-1 SAR (synthetic aperture radar) imagery
- Sink detection from ArcticDEM digital elevation models
+Post-processing of these classifications is performed using the GrIML post-processing Python package, including raster-to-vector conversion, filtering, merging, metadata population, and statistical analysis.
## Project links
diff --git a/src/griml.egg-info/PKG-INFO b/src/griml.egg-info/PKG-INFO
deleted file mode 100644
index f83ffdc..0000000
--- a/src/griml.egg-info/PKG-INFO
+++ /dev/null
@@ -1,76 +0,0 @@
-Metadata-Version: 2.1
-Name: griml
-Version: 0.0.1
-Summary: A workflow for classifying lakes from satellite imagery and compiling lake inventories
-Home-page: https://github.com/PennyHow/GrIML
-Author: Penelope How
-Author-email: pho@geus.dk
-License: UNKNOWN
-Project-URL: Bug Tracker, https://github.com/PennyHow/GrIML/issues
-Keywords: glaciology ice lake ESA
-Platform: UNKNOWN
-Classifier: Programming Language :: Python :: 3
-Classifier: License :: OSI Approved :: MIT License
-Classifier: Development Status :: 4 - Beta
-Classifier: Intended Audience :: Science/Research
-Classifier: Natural Language :: English
-Classifier: Topic :: Scientific/Engineering
-Classifier: Operating System :: OS Independent
-Requires-Python: >=3.6
-Description-Content-Type: text/markdown
-License-File: LICENSE
-
-# Investigating Greenland's ice marginal lakes under a changing climate (GrIML)
-
-[](https://griml.readthedocs.io/en/latest/?badge=latest)
-
-A repository for all project-related materials, funded under the ESA Living Planet Fellowship.
-
-**Project aim:** To examine ice marginal lake changes across Greenland using a multi-method and multi-sensor remote sensing approach, refined with in situ validation.
-
-## Background
-
- -
-Sea level is predicted to rise drastically by 2100, with significant contribution from the melting of the Greenland Ice Sheet (GrIS). In these predictions, melt runoff is assumed to contribute directly to sea level change, with little consideration for meltwater storage at the terrestrial margin of the ice sheet; such as ice marginal lakes.
-
-In 2017, 3347 ice marginal lakes were identified in Greenland along the ice margin (How et al., 2021, see map figure for all mapped lakes). Globally, these ice marginal lakes hold up to 0.43 mm of sea level equivalent, which could have a marked impact on future predictions (Shugar et al., 2021). Therefore, they need to be monitored to understand how changes in ice marginal lake water storage affect melt contribution, and how their dynamics evolve under a changing climate.
-
-**GrIML** proposes to examine ice marginal lake changes across Greenland using a multi-sensor and multi-method remote sensing approach to better address their influence on sea level contribution forecasting.
-
-1. Greenland-wide inventories of ice marginal lakes will be generated for selected years during the satellite era, building upon established classification methods in a unified cloud processing workflow
-
-2. Detailed time-series analysis will be conducted on chosen ice marginal lakes to assess changes in their flooding dynamics; focusing on lakes with societal and scientific importance
-
-3. The findings from this work will be validated against in situ observations - namely hydrological measurements and terrestrial time-lapse images - to evaluate whether the remote sensing workflow adequately captures ice marginal lake dynamics
-
-
-## Methodology
-
-Ice marginal lakes will be detected using a remote sensing approach, based on offline workflows developed within the ESA Glaciers CCI (Option 6, An Inventory of Ice-Marginal Lakes in Greenland) (see workflow below). Lake extents were defined through a multi-sensor approach, using multi-spectral indices classification from Sentinel-2 optical imagery, backscatter classification from Sentinel-1 SAR (synthetic aperture radar) imagery, and sink detection from ArcticDEM digital elevation models (How et al., 2021).
-
-
-
-Sea level is predicted to rise drastically by 2100, with significant contribution from the melting of the Greenland Ice Sheet (GrIS). In these predictions, melt runoff is assumed to contribute directly to sea level change, with little consideration for meltwater storage at the terrestrial margin of the ice sheet; such as ice marginal lakes.
-
-In 2017, 3347 ice marginal lakes were identified in Greenland along the ice margin (How et al., 2021, see map figure for all mapped lakes). Globally, these ice marginal lakes hold up to 0.43 mm of sea level equivalent, which could have a marked impact on future predictions (Shugar et al., 2021). Therefore, they need to be monitored to understand how changes in ice marginal lake water storage affect melt contribution, and how their dynamics evolve under a changing climate.
-
-**GrIML** proposes to examine ice marginal lake changes across Greenland using a multi-sensor and multi-method remote sensing approach to better address their influence on sea level contribution forecasting.
-
-1. Greenland-wide inventories of ice marginal lakes will be generated for selected years during the satellite era, building upon established classification methods in a unified cloud processing workflow
-
-2. Detailed time-series analysis will be conducted on chosen ice marginal lakes to assess changes in their flooding dynamics; focusing on lakes with societal and scientific importance
-
-3. The findings from this work will be validated against in situ observations - namely hydrological measurements and terrestrial time-lapse images - to evaluate whether the remote sensing workflow adequately captures ice marginal lake dynamics
-
-
-## Methodology
-
-Ice marginal lakes will be detected using a remote sensing approach, based on offline workflows developed within the ESA Glaciers CCI (Option 6, An Inventory of Ice-Marginal Lakes in Greenland) (see workflow below). Lake extents were defined through a multi-sensor approach, using multi-spectral indices classification from Sentinel-2 optical imagery, backscatter classification from Sentinel-1 SAR (synthetic aperture radar) imagery, and sink detection from ArcticDEM digital elevation models (How et al., 2021).
-
- -
-The intent in GrIML is to build upon this pre-existing workflow with new and innovative solutions to form a unified and automated processing chain, with the option to integrate it into a cloud processing platform for efficient big data analysis.
-
-These developments will alleviate the current challenges associated with data-heavy processing (i.e. multi-sensor integration and data retrieval), and ensure detection accuracy with a merged and collated set of established methodologies. These developments will include:
-
-1. Incorporation of the multi-spectral indices classification, backscatter classification, and sink detection methods into one unified processing chain
-
-2. Integration of image analysis from additional sensors into the processing chain, such as Landsat 4-8
-
-3. Automisation of post-processing filtering to remove detached water bodies and misclassifications, using the already-existing 2017 inventory (How et al., 2021) as a training dataset to automatically match and retain existing lakes
-
-4. Adaptation of the workflow for easy transference between offline and cloud processing platforms
-
-## Links
-
-- ESA project outline and fellow information
-
-- Information about the ESA Living Planet Fellowship
-
-- GrIML project description
-
-- 2017 ice marginal lake inventory Scientific Reports paper and dataset
-
-
diff --git a/src/griml.egg-info/SOURCES.txt b/src/griml.egg-info/SOURCES.txt
deleted file mode 100644
index b38c4c2..0000000
--- a/src/griml.egg-info/SOURCES.txt
+++ /dev/null
@@ -1,15 +0,0 @@
-LICENSE
-README.md
-setup.py
-src/griml/__init__.py
-src/griml/dem.py
-src/griml/lake.py
-src/griml/process.py
-src/griml/retrieve.py
-src/griml/sar.py
-src/griml/vis.py
-src/griml.egg-info/PKG-INFO
-src/griml.egg-info/SOURCES.txt
-src/griml.egg-info/dependency_links.txt
-src/griml.egg-info/requires.txt
-src/griml.egg-info/top_level.txt
\ No newline at end of file
diff --git a/src/griml.egg-info/dependency_links.txt b/src/griml.egg-info/dependency_links.txt
deleted file mode 100644
index 8b13789..0000000
--- a/src/griml.egg-info/dependency_links.txt
+++ /dev/null
@@ -1 +0,0 @@
-
diff --git a/src/griml.egg-info/requires.txt b/src/griml.egg-info/requires.txt
deleted file mode 100644
index fe00572..0000000
--- a/src/griml.egg-info/requires.txt
+++ /dev/null
@@ -1,6 +0,0 @@
-earthengine-api
-geopandas
-numpy
-pandas
-scipy
-Shapely
diff --git a/src/griml.egg-info/top_level.txt b/src/griml.egg-info/top_level.txt
deleted file mode 100644
index ba4ef6c..0000000
--- a/src/griml.egg-info/top_level.txt
+++ /dev/null
@@ -1 +0,0 @@
-griml
diff --git a/src/griml/examples/getInventory_funcs.py b/src/griml/examples/getInventory_funcs.py
deleted file mode 100644
index 0dea76c..0000000
--- a/src/griml/examples/getInventory_funcs.py
+++ /dev/null
@@ -1,406 +0,0 @@
-"""
-GrIML processing module playground (experimental)
-
-@author: Penelope How
-"""
-
-import ee, time, sys
-from time import gmtime, strftime
-import requests, zipfile, io, urllib
-from urllib.error import HTTPError
-
-import numpy as np
-from shapely.ops import split
-from shapely.geometry import Polygon, LineString, MultiPolygon
-import geopandas as gpd
-import multiprocessing as mp
-
-sys.path.append('../')
-from retrieve import getScenes, getScene, getInt, getSmooth, getMosaic, \
- getMean, maskImage, splitBBox, getVectors, extractFeatures, getFeatures, \
- getFeaturesSplit
-from sar import filterSARscenes, classifySARimage
-from vis import filterS2scenes, maskS2clouds, renameS2scenes, \
- resampleS2scenes, classifyVISimage, filterLSscenes, maskL8clouds, \
- renameL8scenes
-from dem import getElevation, getSlope, getSinks
-from lake import compileFeatures, assignID, assignSources, assignNames
-
-start=time.time()
-
-#------------------------------------------------------------------------------
-
-# Set AOI
-
-aoi1 = [-50.94, 67.98] # SW test region
-aoi2 = [-49.66, 67.58]
-
-# aoi1 = [-54.68242187500001,68.8415979398901]
-# aoi2 = [-47.21171875000001,60.62185574504481] # SW region
-
-# aoi1 = [-10.297656250000014,59.03190534154833] # All Greenland
-# aoi2 = [-74.98515625000002,83.9867103173338]
-
-print('Loading bounding box')
-aoi = '/home/pho/Desktop/python_workspace/GrIML/other/datasets/aoi/test_mask_4326.shp'
-aoi = gpd.read_file(aoi)
-aoi_poly = aoi.geometry.iloc[0]
-xmin, ymin, xmax, ymax = aoi_poly.bounds
-
-# Set AOI box splitter parameters
-wh=0.3 # Window height
-ww=0.3 # Window width
-oh = 0.05 # Overlap height
-ow = 0.05 # Overlap width
-
-# Set date range
-date1='2017-07-01'
-date2='2017-08-31'
-
-# Set maximum cloud cover
-cloud=50
-
-# Set mask location
-ice_mask = 'users/pennyruthhow/GIMP_iceMask_edit_buffer7km'
-
-# Set output projection
-proj = 'EPSG:3413' # Polar stereographic
-
-
-#--------------------------- Initialise GEE -------------------------------
-
-ee.Initialize()
-print('EE initialized')
-
-# Set the Area of Interest (AOI) through box coordinates
-box = ee.Geometry.Rectangle([xmin, ymin, xmax, ymax])
-
-# Split box into grid for exporting
-bbox = splitBBox(aoi_poly, wh, ww, oh, ow)
-print('grid completed')
-# # Remove boxes that don't overlap with AOI polygon
-# grid = [Polygon([[g[0],g[1]], [g[0],g[3]], [g[2],g[3]],
-# [g[2],g[1]], [g[0],g[1]]]) for g in grid]
-# bbox = [ee.Geometry.Rectangle(min(g.exterior.coords.xy[0]),
-# min(g.exterior.coords.xy[1]),
-# max(g.exterior.coords.xy[0]),
-# max(g.exterior.coords.xy[1])) for g in grid if g.intersects(aoi_poly)]
-
-
-print(f'Computed {len(bbox)} bounding boxes')
-print(f'Total area covered: {aoi_poly.area} sq km')
-
-
-
-
-# bbox = bbox[100:105]
-
-# # Set the Area of Interest (AOI) through box coordinates
-# box = ee.Geometry.Rectangle([aoi1[0], aoi1[1],
-# aoi2[0], aoi2[1]])
-
-# bbox = [ee.Geometry.Rectangle(g[0], g[1], g[2], g[3]) for g in grid]
-
-# print(f'Computed {len(bbox)} bounding boxes from {aoi1}, {aoi2}')
-# print(f'Total area covered: {box.area().getInfo()/10**6} sq km')
-
-
-#--------------------- Get basic ocean mask ---------------------------------
-
-# Retrieve ice-ocean mask
-img = ee.Image("OSU/GIMP/2000_ICE_OCEAN_MASK")
-ocean_mask = img.select('ocean_mask').eq(0)
-# ice_mask = img.select('ice_mask').eq(0)
-
-# imask0 = ice_mask.reduceToVectors(geometryType='polygon', labelProperty='v', scale=10, bestEffort=True)
-# def _mapBuffer(feature):
-# return feature.buffer(-10000)
-# imask01 = imask0.map(_mapBuffer)
-# imask0 = imask01.reduceToImage(properties=['v'], reducer=ee.Reducer.mean())
-
-# sys.exit(1)
-
-# # Construct vector features
-# def getFeaturesSplit(image, bbox):
-# features=[]
-# for b in range(len(bbox)):
-# print(f'Fetching vectors from bbox {b}...')
-# v = getVectors(image, 10, bbox[b])
-# features.append(extractFeatures(v))
-# features = [val for sublist in features for val in sublist]
-# print(f'{len(features)} vector features identified')
-# return features
-
-# def getFeatures(image, bbox):
-# features=[]
-# v = getVectors(image, 10, bbox)
-# features = extractFeatures(v)
-# print(f'{len(features)} vector features identified')
-# return features
-
-# def getParallelFeatures1(image, bbox):
-# # Parallel process vector generation
-# pool = mp.Pool(mp.cpu_count())
-# v = [pool.apply(getVectors, args=(image, 10, bbox[b])) for b in range(len(bbox))]
-# pool.close()
-
-# # Parallel process feature extraction
-# pool = mp.Pool(mp.cpu_count())
-# features = pool.map(extractFeatures, [row for row in v])
-# pool.close()
-
-# features = [val for sublist in features for val in sublist]
-# print(f'{len(features)} vector features identified')
-# return features
-
-# def getDownload(v):
-# link = v.getDownloadURL('csv')
-# req = urllib.request.Request(url=link)
-# try:
-# handler = urllib.request.urlopen(req)
-# except HTTPError as e:
-# handler = e.read()
-# lines = []
-# for l in handler:
-# lines.append(str(l))
-# features_dem.append(lines)
-
-#--------------------------- DEM processing -------------------------------
-
-# Set collection
-dem_col = 'UMN/PGC/ArcticDEM/V3/2m_mosaic'
-
-# Get image collection
-scenes = getScene(dem_col, box)
-print(f'\nScenes gathered from {dem_col}')
-
-# Get elevation and slope
-elevation = getElevation(scenes)
-slope = getSlope(elevation)
-
-elevation = getSmooth(elevation, 110)
-
-# Mask out ocean pixels
-elevation = maskImage(elevation, ocean_mask)
-
-# Get sinks
-elevation = getInt(elevation)
-sinks = getSinks(elevation, 10, 50)
-
-# Remove speckle with smoothing
-sinks = getSmooth(sinks, 50).rename('dem_sinks')
-sinks = getInt(sinks)
-print(f'{dem_col} scenes classified')
-print('Band names: ' + str(sinks.bandNames().getInfo()))
-
-try:
- features_dem = getFeatures(sinks, 10, box)
-except:
- features_dem = getFeaturesSplit(sinks, 10, bbox)
-
-
-#--------------------------- SAR processing -------------------------------
-
-# Set collection
-sar_col = 'COPERNICUS/S1_GRD'
-
-# Get image collection
-scenes = getScenes(sar_col, date1, date2, box)
-if scenes.size().getInfo() > 0:
- scenes = filterSARscenes(scenes)
- print(f'\nScenes gathered from {sar_col}')
- print('Total number of scenes: ', scenes.size().getInfo())
- print('Number of bands per scene: '
- + str(len(scenes.first().bandNames().getInfo())))
- print('Band names: ' + str(scenes.first().bandNames().getInfo()))
-
- # Get average
- aver = getMosaic(scenes, 'HH')
-
- # Mask out ocean pixels
- aver = maskImage(aver, ocean_mask)
-
- # Classify water from SAR average image
- water_sar = classifySARimage(aver, -20, 'HH', 50)
- print(f'{sar_col} scenes classified')
- print('Band names: ' + str(water_sar.bandNames().getInfo()))
-
- try:
- features_sar = getFeatures(water_sar, 10, box)
- except:
- features_sar = getFeaturesSplit(water_sar, 10, bbox)
-
-else:
- features_sar = None
- print(f'\nNo {sar_col} scenes identified between {date1} and {date2}')
-
-#------------------------- VIS processing (S2) -----------------------------
-
-# Set collection
-vis_col1 = "COPERNICUS/S2"
-
-# Get image collection
-scenes = getScenes(vis_col1, date1, date2, box)
-if scenes.size().getInfo() > 0:
- scenes = filterS2scenes(scenes, cloud)
- print(f'\nScenes gathered from {vis_col1}')
- print('Total number of scenes: ', scenes.size().getInfo())
- print('Number of bands per scene: '
- + str(len(scenes.first().bandNames().getInfo())))
- print('Band names: ' + str(scenes.first().bandNames().getInfo()))
-
- # Mask scenes for clouds
- scenes = maskS2clouds(scenes)
- # ee.Terrain.hillshade(image, azimuth, elevation)
-
- # Get average of spectific bands
- scenes = renameS2scenes(scenes)
- scenes = resampleS2scenes(scenes)
- aver = getMean(scenes, ['blue','green','red','vnir','swir1_1','swir2_1'])
- print('Scenes resampled and mosiacked')
- print('Band names: ' + str(aver.bandNames().getInfo()))
-
- # Classify water from VIS average image, and mask out ocean pixels
- water_s2 = classifyVISimage(aver)
- water_s2 = maskImage(water_s2, ocean_mask)
- print(f'{vis_col1} scenes classified')
- print('Band names: ' + str(water_s2.bandNames().getInfo()))
-
- try:
- features_s2 = getFeatures(water_s2, 10, box)
- except:
- features_s2 = getFeaturesSplit(water_s2, 10, bbox)
-else:
- features_s2 = None
- print(f'\nNo {vis_col1} scenes identified between {date1} and {date2}')
-
-#----------------------- VIS processing (Landsat 8) -----------------------
-
-# Set collection
-vis_col2 = "LANDSAT/LC08/C01/T1_TOA"
-
-# Get image collection
-scenes = getScenes(vis_col2, date1, date2, box)
-if scenes.size().getInfo() > 0:
- scenes = filterLSscenes(scenes, cloud)
- print(f'\nScenes gathered from {vis_col2}')
- print('Total number of scenes: ', scenes.size().getInfo())
- print('Number of bands per scene: '
- + str(len(scenes.first().bandNames().getInfo())))
- print('Band names: ' + str(scenes.first().bandNames().getInfo()))
-
- # Mask scenes for clouds
- scenes = maskL8clouds(scenes)
- # ee.Terrain.hillshade(image, azimuth, elevation)
-
- # Get average of spectific bands
- scenes = renameL8scenes(scenes)
- aver = getMean(scenes, ['blue','green','red','vnir','swir1','swir2'])
- print('Scenes resampled and mosiacked')
- print('Band names: ' + str(aver.bandNames().getInfo()))
-
- # Classify water from VIS average image, and mask out ocean pixels
- ndwi = aver.normalizedDifference(['green_mean', 'vnir_mean']).rename('ndwi')
- mndwi = aver.normalizedDifference(['green_mean','swir1_mean']).rename('mndwi')
- aweish = aver.expression('BLUE + 2.5 * GREEN - 1.5 * (VNIR + SWIR1) - 0.25 * SWIR2',
- {'BLUE' : aver.select('blue_mean'),
- 'GREEN' : aver.select('green_mean'),
- 'SWIR1' : aver.select('swir1_mean'),
- 'VNIR' : aver.select('vnir_mean'),
- 'SWIR2' : aver.select('swir2_mean')}).rename('aweish')
- aweinsh = aver.expression('4.0 * (GREEN - SWIR1) - (0.25 * VNIR + 2.75 * SWIR2)',
- {'GREEN' : aver.select('green_mean'),
- 'SWIR1' : aver.select('swir1_mean'),
- 'VNIR' : aver.select('vnir_mean'),
- 'SWIR2' : aver.select('swir2_mean')}).rename('aweinsh')
- bright = aver.expression('(RED + GREEN + BLUE) / 3',
- {'BLUE' : aver.select('blue_mean'),
- 'GREEN' : aver.select('green_mean'),
- 'RED' : aver.select('red_mean')}).rename('bright')
-
- aver = aver.addBands([ndwi, mndwi, aweish, aweinsh, bright])
- classified = aver.expression("(BRIGHT > 5000) ? 0"
- ": (NDWI > 0.3) ? 1 "
- ": (MNDWI < 0.1) ? 0 "
- ": (AWEISH < 2000) ? 0"
- ": (AWEISH > 5000) ? 0"
- ": (AWEINSH < 4000) ? 0"
- ": (AWEINSH > 6000) ? 0"
- ": 1",
- {'NDWI' : aver.select('ndwi'),
- 'MNDWI' : aver.select('mndwi'),
- 'AWEISH' : aver.select('aweish'),
- 'AWEINSH' : aver.select('aweinsh'),
- 'BRIGHT' : aver.select('bright')}).rename('water')
- water_ls8 = classified.updateMask(classified)
- water_ls8 = maskImage(water_ls8, ocean_mask)
- print(f'{vis_col2} scenes classified')
- print('Band names: ' + str(water_ls8.bandNames().getInfo()))
-
- try:
- features_ls8 = getFeatures(water_ls8, 10, box)
- except:
- features_ls8 = getFeaturesSplit(water_ls8, 10, bbox)
-
-else:
- features_ls8 = None
- print(f'\nNo {vis_col2} scenes identified between {date1} and {date2}')
-
-
-#------------------------ Compile geodatabase -----------------------------
-
-iml = compileFeatures([features_s2, features_ls8, features_sar, features_dem],
- ['VIS','VIS','SAR','DEM'],
- [vis_col1, vis_col2, sar_col, dem_col],
- [date1, date2])
-print(f'\nCompiled {iml.shape[0]} geodatabase features')
-
-# Reproject geometries
-iml = iml.to_crs(proj)
-print(f'{iml.shape[0]} unfiltered features')
-
-iml.to_file(f"out/iiml_{date1}_{date2}_{aoi1[0]}_{aoi1[1]}_unfiltered.shp")
-
-
-#-------------------- Filter database by ice margin -----------------------
-
-# Load margin and add buffer
-margin = gpd.read_file("../../../other/datasets/ice_margin/gimp_icemask_line_polstereo.shp")
-margin_buff = margin.buffer(500)
-margin_buff = gpd.GeoDataFrame(geometry=margin_buff, crs=margin.crs)
-
-# Perform spatial join
-iml = gpd.sjoin(iml, margin_buff, how='left')
-iml = iml[iml['index_right']==0]
-iml = iml.drop(columns='index_right')
-print(f'{iml.shape[0]} features within 500 m of margin')
-
-# Filter lakes by area
-iml['area_sqkm'] = iml['geometry'].area/10**6
-iml['length_km'] = iml['geometry'].length/1000
-iml = iml[(iml.area_sqkm >= 0.05)]
-
-# Calculate geometry info
-iml.reset_index(inplace=True, drop=True)
-print(f'{iml.shape[0]} features over 0.05 sq km')
-iml.to_file(f"out/iiml_{date1}_{date2}_{aoi1[0]}_{aoi1[1]}_filtered.shp")
-
-#------------------------- Populate metadata ------------------------------
-
-iml = assignID(iml)
-iml = assignSources(iml)
-
-names = gpd.read_file('../../../other/datasets/placenames/oqaasileriffik_placenames.shp')
-iml = assignNames(iml, names)
-
-#----------------------- Write data to shapefile --------------------------
-
-iml.to_file(f"/home/pho/Desktop/python_workspace/GrIML/other/out/iiml_{date1}_{date2}_{aoi1[0]}_{aoi1[1]}_final.shp")
-print(f'Saved to out/iiml_{date1}_{date2}_{aoi1[0]}_{aoi1[1]}_final.shp')
-
-# Plot lakes
-iml.plot(figsize=(10, 10), alpha=0.5, edgecolor='k')
-
-print('\nFinished')
-end=time.time()
-print('Total run time: ' + strftime("%H:%M:%S", gmtime(round(end-start))))
diff --git a/src/griml/examples/getInventory_obj.py b/src/griml/examples/getInventory_obj.py
deleted file mode 100644
index f8b507c..0000000
--- a/src/griml/examples/getInventory_obj.py
+++ /dev/null
@@ -1,182 +0,0 @@
-"""
-GrIML processing module playground (experimental)
-
-@author: Penelope How
-"""
-
-import time,sys
-from time import gmtime, strftime
-import geopandas as gpd
-
-try:
- sys.path.append('../')
- from process import gee
- from lake import dissolvePolygons, compileFeatures, assignID, assignSources, assignNames
-except:
- from griml.process import gee
- from griml.lake import dissolvePolygons, compileFeatures, assignID, \
- assignSources, assignNames
-
-start=time.time()
-
-#------------------------------------------------------------------------------
-
-# Set AOI box splitter parameters
-wh=0.3 # Window height
-ww=0.3 # Window width
-oh = 0.05 # Overlap height
-ow = 0.05 # Overlap width
-
-# Set date range
-date1='2017-07-01'
-date2='2017-08-31'
-
-# Set output projection
-proj = 'EPSG:3413' # Polar stereographic
-
-# Set AOI
-# aoi = '/home/pho/python_workspace/GrIML/other/datasets/aoi/AOI_mask_split2.shp'
-aoi = '/home/pho/python_workspace/GrIML/other/datasets/aoi/test_mask_4326.shp'
-aoi = gpd.read_file(aoi)#.to_crs('EPSG:4326')
-
-# Load ice margin with buffer
-print('Preparing ice margin buffer...')
-margin_buff = gpd.read_file("/home/pho/python_workspace/GrIML/other/datasets/ice_margin/gimp_icemask_line_polstereo_simple_buffer.shp")
-# margin_buff = margin.buffer(500)
-# margin_buff = gpd.GeoDataFrame(geometry=margin_buff, crs=margin.crs)
-
-# Load name database
-print('Loading placename database...')
-names = gpd.read_file('/home/pho/python_workspace/GrIML/other/datasets/placenames/oqaasileriffik_placenames.shp')
-
-
-#--------------------------- Initialise GEE -------------------------------
-
-parameters = [#{'collection' : 'UMN/PGC/ArcticDEM/V3/2m_mosaic',
- #'smooth' : 100, 'fill' : 100, 'kernel' : 100, 'speckle' : 50},
- # {'collection' : 'UMN/PGC/ArcticDEM/V3/2m',
- # 'smooth' : 100, 'fill' : 100, 'kernel' : 100, 'speckle' : 50},
- {'collection' : 'COPERNICUS/S1_GRD',
- 'polar' : 'HH', 'threshold' : -20, 'smooth' : 50},
- #{'collection' : 'COPERNICUS/S2', `
- #'cloud' : 20},
- {'collection' : 'LANDSAT/LC08/C01/T1_TOA',
- 'cloud' : 20}]
- # {'collection' : 'LANDSAT/LE07/C02/T1_TOA',
- # 'cloud' : 50}]
-
-
-
-for i in range(len(aoi.index)):
- print(f'\nCommencing processing of {list(aoi["catch"])[i]} catchment area')
- aoi_poly = aoi.geometry.iloc[i]
- xmin, ymin, xmax, ymax = aoi_poly.bounds
-
- print('Clipping margin to catchment area...')
- aoi_poly_proj = gpd.GeoDataFrame(geometry=[aoi_poly], crs=aoi.crs).to_crs(proj)
- margin_clip = gpd.clip(margin_buff, aoi_poly_proj)
-
- print('Conducting classifications...')
- proc = gee([date1, date2], aoi_poly, parameters, [wh, ww, oh, ow], True)
-
- print('Processing...')
- water = proc.processAll()
-
- print('Retrieving...')
- features = proc.retrieveAll(water)
-
- # Filter features
- filtered=[]
- for f in features:
- print('Filtering features...')
-
- # Remove duplicates
- print('Dissolving polygons...')
- gdf = gpd.GeoDataFrame(geometry=f, crs='EPSG:4326')
- gdf = dissolvePolygons(gdf)
-
- # Remove lakes below size threshold
- print('Filtering by size...')
- gdf = gdf.set_crs('EPSG:4326')
- gdf = gdf.to_crs(proj)
- gdf['area_sqkm'] = gdf['geometry'].area/10**6
- gdf = gdf[(gdf.area_sqkm >= 0.05)]
-
- # # Remove lakes outside of margin boundary
- # print('Filtering by bounds...')
- # gdf = gpd.sjoin(gdf, margin_clip, how='left')
- # gdf = gdf[gdf['index_right']==0]
- # gdf = gdf.drop(columns='index_right')
- filtered.append(gdf.geometry)
-
-
-#------------------------ Compile geodatabase -----------------------------
-# lakes=[]
-# for f in features:
-# gdf = gpd.GeoDataFrame(geometry=f, crs='EPSG:4326')
-# gdf = dissolvePolygons(gdf)
-# gdf['area'] = gdf['geometry'].area/10**6
-# gdf = gdf[(gdf.area >= 0.05)]
-
-# gdf = gpd.sjoin(gdf, margin_buff, how='left')
-# gdf = gdf[gdf['index_right']==0]
-# gdf = gdf.drop(columns='index_right')
-# lakes.append(list(gdf['geometry']))
-
- # Compile all features
- iml = compileFeatures(filtered,
- ['DEM', 'SAR', 'VIS'],
- ['UMN/PGC/ArcticDEM/V3/2m_mosaic', 'COPERNICUS/S1_GRD',
- 'LANDSAT/LC08/C01/T1_TOA'],
- [date1, date2], proj)
- print(f'\nCompiled {iml.shape[0]} geodatabase features')
-
- # Calculate geometry attributes
- iml['area_sqkm'] = iml['geometry'].area/10**6
- iml['length_km'] = iml['geometry'].length/1000
- iml = iml[(iml.area_sqkm >= 0.05)]
- iml['catch'] = str(list(aoi["catch"])[i])
-
- # Calculate geometry info
- iml.reset_index(inplace=True, drop=True)
-
- print(f'{iml.shape[0]} filtered features')
-
- iml.to_file(f'out/iiml_{date1}_{date2}_{list(aoi["catch"])[i]}_filtered.shp')
-
-
-# #-------------------- Filter database by ice margin -----------------------
-
-# # Perform spatial join
-# aoi_poly_proj = aoi_poly.to_crs(proj)
-# margin_clip = gpd.clip(margin_buff, aoi_poly_proj)
-
-# iml = gpd.sjoin(iml, margin_clip, how='left')
-# iml = iml[iml['index_right']==0]
-# iml = iml.drop(columns='index_right')
-# print(f'{iml.shape[0]} features within 500 m of margin')
-
-# # Calculate geometry info
-# iml.reset_index(inplace=True, drop=True)
-# print(f'{iml.shape[0]} features over 0.05 sq km')
-# iml.to_file(f'out/iiml_{date1}_{date2}_{list(aoi["catch"])[i]}_filtered.shp')
-
-
-#-------------------- Populate metadata and write to file -----------------
-
-# iml = assignID(iml)
- # iml = assignSources(iml)
-
- # iml = assignNames(iml, names)
-
- # iml.to_file(f'out/iiml_{date1}_{date2}_{list(aoi["catch"])[i]}_final.shp')
- # print(f'Saved to out/iiml_{date1}_{date2}_{list(aoi["catch"])[i]}_final.shp')
-
- # # Plot lakes
- # iml.plot(figsize=(10, 10), alpha=0.5, edgecolor='k')
-
-
-#------------------------------------------------------------------------------
-print('\nFinished')
-end=time.time()
-print('Total run time: ' + strftime("%H:%M:%S", gmtime(round(end-start))))
diff --git a/src/griml/metadata/__init__.py b/src/griml/metadata/__init__.py
index 3d16982..7e1e3d8 100644
--- a/src/griml/metadata/__init__.py
+++ b/src/griml/metadata/__init__.py
@@ -1,5 +1,6 @@
from griml.metadata.assign_certainty import *
from griml.metadata.assign_id import *
from griml.metadata.assign_names import *
+from griml.metadata.assign_regions import *
from griml.metadata.assign_sources import *
from griml.metadata.add_metadata import *
diff --git a/src/griml/metadata/add_metadata.py b/src/griml/metadata/add_metadata.py
index 2f15ee2..50e6d9a 100644
--- a/src/griml/metadata/add_metadata.py
+++ b/src/griml/metadata/add_metadata.py
@@ -1,30 +1,17 @@
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
-from griml.metadata import assign_id, assign_sources, assign_certainty, \
- assign_names
from griml.load import load
+from griml.metadata import assign_id, assign_sources, assign_certainty, \
+ assign_names, assign_regions
import geopandas as gpd
-def add_metadata(iml_file, names_file, outfile=None):
- '''Add metadata to collection of features
-
- Parameters
- ----------
- iml_file : str, geopandas.dataframe.DataFrame
- Filepath or geopandas.dataframe.DataFrame object to assign metadata to
-
- Returns
- -------
- iml: geopandas.dataframe.GeoDataFrame
- Geodataframe with metadata
- '''
-
- # Load files
- iml = load(iml_file)
- names = load(names_file)
+def add_metadata(iml, names, regions, outfile=None):
+
+ iml = load(iml)
+ names = load(names)
+ regions = load(regions)
- # Perform metadata steps
print('Assigning ID...')
iml = assign_id(iml)
@@ -35,24 +22,27 @@ def add_metadata(iml_file, names_file, outfile=None):
n = ['S1','S2','ARCTICDEM']
scores = [0.298, 0.398, 0.304]
iml = assign_certainty(iml, n, scores)
+
+ print('Assigning regions...')
+ iml = assign_regions(iml, regions)
print('Assigning placenames...')
iml = assign_names(iml, names)
- # Save to file if given
- if outfile is not None:
+ if outfile:
print('Saving file...')
iml.to_file(outfile)
- print('Saved to '+str(outfile))
-
- return iml
+ print('Saved to '+str(outfile)+"_metadata.shp")
if __name__ == "__main__":
- infile1 = '../test/test_merge_2.shp'
- infile2 = '../test/test_placenames.shp'
- add_metadata(infile1, infile2)
+ # indir = "/home/pho/python_workspace/GrIML/other/iml_2017/merged_vectors/griml_2017_inventory.shp"
+ indir = "/home/pho/python_workspace/GrIML/other/iml_2017/inspected_vectors/lakes_all-0000000000-0000037888_filtered.shp"
+
+
+ infile_names = '/home/pho/python_workspace/GrIML/other/datasets/placenames/oqaasileriffik_placenames.shp'
+ infile_regions = '/home/pho/python_workspace/GrIML/other/datasets/drainage_basins/greenland_basins_polarstereo.shp'
+
- iml = gpd.read_file(infile1)
- names = gpd.read_file(infile2)
- add_metadata(iml, names)
\ No newline at end of file
+ outfile = "/home/pho/python_workspace/GrIML/other/iml_2017/metadata_vectors/griml_2017_inventory_final.shp"
+ add_metadata(indir, infile_names, infile_regions, outfile)
diff --git a/src/griml/metadata/assign_names.py b/src/griml/metadata/assign_names.py
index 9bd294f..c151a48 100644
--- a/src/griml/metadata/assign_names.py
+++ b/src/griml/metadata/assign_names.py
@@ -1,7 +1,18 @@
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
-def assign_names(gdf, gdf_names, distance=500.0):
+import itertools
+from operator import itemgetter
+
+import geopandas as gpd
+import numpy as np
+import pandas as pd
+
+from scipy.spatial import cKDTree
+from shapely.geometry import Point, LineString, Polygon
+from griml.load import load
+
+def assign_names(gdf, gdf_names):
'''Assign placenames to geodataframe geometries based on names in another
geodataframe point geometries
@@ -19,62 +30,41 @@ def assign_names(gdf, gdf_names, distance=500.0):
gdf : pandas.GeoDataFrame
Vectors with assigned IDs
'''
- placenames = _compile_names(gdf_names)
+
+ # Load geodataframes
+ gdf1 = load(gdf)
+ gdf2 = load(gdf_names)
+
+ # Compile placenames into new dataframe
+ names = _compile_names(gdf2)
+ placenames = gpd.GeoDataFrame({"geometry": list(gdf2['geometry']),
+ "placename": names})
- lakename=[]
- for i,v in gdf.iterrows():
-
- geom = v['geometry']
-
- polynames=[]
- for pt in range(len(placenames)):
- if geom.contains(gdf_names['geometry'][pt]) == True:
- polynames.append(placenames[pt])
-
- if len(polynames)==0:
- for pt in range(len(placenames)):
- if gdf_names['geometry'][pt].distance(geom) < distance:
- polynames.append(placenames[pt])
- lakename.append(polynames)
-
- elif len(polynames)==1:
- lakename.append(polynames)
-
- else:
- out=[]
- for p in polynames:
- out.append(p)
- lakename.append(out)
-
- lakeid = gdf['unique_id'].tolist()
- lakename2 = []
+ # Assign names based on proximity
+ a = _get_nearest_point(gdf1, placenames)
+ return a
+
+
+def _get_nearest_point(gdA, gdB, distance=500.0):
+ '''Return properties of nearest point in Y to geometry in X'''
+ nA = np.array(list(gdA.geometry.centroid.apply(lambda x: (x.x, x.y))))
+ nB = np.array(list(gdB.geometry.apply(lambda x: (x.x, x.y))))
+
+ btree = cKDTree(nB)
+ dist, idx = btree.query(nA, k=1)
+ gdB_nearest = gdB.iloc[idx].drop(columns="geometry").reset_index(drop=True)
+ gdf = pd.concat(
+ [
+ gdA.reset_index(drop=True),
+ gdB_nearest,
+ pd.Series(dist, name='dist')
+ ],
+ axis=1)
- for x in gdf.index:
- indx = _get_indices(lakeid, x)
- findname=[]
- for l in indx:
- if len(lakename[l])!=0:
- findname.append(lakename[l])
-
- for i in range(len(indx)):
- if len(findname)==0:
- lakename2.append('')
-
- else:
- unique = set(findname[0])
- unique = list(unique)
-
- if len(unique)==1:
- lakename2.append(findname[0][0])
-
- else:
- out2 = ', '
- out2 = out2.join(unique)
- lakename2.append(out2)
- gdf['placename'] = lakename2
+ gdf.loc[gdf['dist']>=distance, 'placename'] = 'Unknown'
+ gdf = gdf.drop(columns=['dist'])
return gdf
-
def _get_indices(mylist, value):
'''Get indices for value in list'''
return[i for i, x in enumerate(mylist) if x==value]
@@ -95,3 +85,14 @@ def _compile_names(gdf):
else:
placenames.append(None)
return placenames
+
+
+if __name__ == "__main__":
+
+ # Define file attributes
+ infile = '/home/pho/Desktop/python_workspace/GrIML/src/griml/test/test_merge_1.shp'
+ names = '/home/pho/Desktop/python_workspace/GrIML/other/datasets/placenames/oqaasileriffik_placenames_polarstereo.shp'
+
+
+ # Perform conversion
+ a = assign_names(infile, names)
\ No newline at end of file
diff --git a/src/griml/metadata/assign_region.py b/src/griml/metadata/assign_region.py
deleted file mode 100644
index 9bd294f..0000000
--- a/src/griml/metadata/assign_region.py
+++ /dev/null
@@ -1,97 +0,0 @@
-#!/usr/bin/env python3
-# -*- coding: utf-8 -*-
-
-def assign_names(gdf, gdf_names, distance=500.0):
- '''Assign placenames to geodataframe geometries based on names in another
- geodataframe point geometries
-
- Parameters
- ----------
- gdf : pandas.GeoDataFrame
- Vectors to assign uncertainty to
- gdf_names : pandas.GeoDataFrame
- Vector geodataframe with placenames
- distance : int
- Distance threshold between a given vector and a placename
-
- Returns
- -------
- gdf : pandas.GeoDataFrame
- Vectors with assigned IDs
- '''
- placenames = _compile_names(gdf_names)
-
- lakename=[]
- for i,v in gdf.iterrows():
-
- geom = v['geometry']
-
- polynames=[]
- for pt in range(len(placenames)):
- if geom.contains(gdf_names['geometry'][pt]) == True:
- polynames.append(placenames[pt])
-
- if len(polynames)==0:
- for pt in range(len(placenames)):
- if gdf_names['geometry'][pt].distance(geom) < distance:
- polynames.append(placenames[pt])
- lakename.append(polynames)
-
- elif len(polynames)==1:
- lakename.append(polynames)
-
- else:
- out=[]
- for p in polynames:
- out.append(p)
- lakename.append(out)
-
- lakeid = gdf['unique_id'].tolist()
- lakename2 = []
-
- for x in gdf.index:
- indx = _get_indices(lakeid, x)
- findname=[]
- for l in indx:
- if len(lakename[l])!=0:
- findname.append(lakename[l])
-
- for i in range(len(indx)):
- if len(findname)==0:
- lakename2.append('')
-
- else:
- unique = set(findname[0])
- unique = list(unique)
-
- if len(unique)==1:
- lakename2.append(findname[0][0])
-
- else:
- out2 = ', '
- out2 = out2.join(unique)
- lakename2.append(out2)
- gdf['placename'] = lakename2
- return gdf
-
-
-def _get_indices(mylist, value):
- '''Get indices for value in list'''
- return[i for i, x in enumerate(mylist) if x==value]
-
-
-def _compile_names(gdf):
- '''Get preferred placenames from placename geodatabase'''
- placenames=[]
- for i,v in gdf.iterrows():
- if v['Ny_grønla'] != None:
- placenames.append(v['Ny_grønla'])
- else:
- if v['Dansk'] != None:
- placenames.append(v['Dansk'])
- else:
- if v['Alternativ'] != None:
- placenames.append(v['Alternativ'])
- else:
- placenames.append(None)
- return placenames
diff --git a/src/griml/metadata/assign_regions.py b/src/griml/metadata/assign_regions.py
new file mode 100644
index 0000000..6458989
--- /dev/null
+++ b/src/griml/metadata/assign_regions.py
@@ -0,0 +1,86 @@
+#!/usr/bin/env python3
+# -*- coding: utf-8 -*-
+
+import itertools
+from operator import itemgetter
+
+import geopandas as gpd
+import numpy as np
+import pandas as pd
+
+from griml.load import load
+from scipy.spatial import cKDTree
+from shapely.geometry import Point, LineString, Polygon
+
+def assign_regions(gdf, gdf_regions, region_name='region'):
+ '''Assign region to geodataframe geometries based on regions in another
+ geodataframe object
+
+ Parameters
+ ----------
+ gdf : pandas.GeoDataFrame
+ Vectors to assign region name to
+ gdf_regions : pandas.GeoDataFrame
+ Vector geodataframe with regions
+
+ Returns
+ -------
+ gdf : pandas.GeoDataFrame
+ Vectors with assigned IDs
+ '''
+
+ # Load geodataframes
+ gdf1 = load(gdf)
+ gdf2 = load(gdf_regions)
+
+ g = _get_nearest_polygon(gdf1, gdf2)
+
+ return g
+
+
+def _get_nearest_polygon(gdfA, gdfB, gdfB_cols=['subregion'], distance=100000.0):
+ '''Return given properties of nearest polygon in Y to geometry in X'''
+
+ A = np.array(list(gdfA.geometry.centroid.apply(lambda x: (x.x, x.y))))
+ # B = [np.array(geom.boundary.coords) for geom in gdfB.geometry.to_list()]
+
+ B=[]
+ for geom in gdfB.geometry.to_list():
+ try:
+ g = np.array(geom.boundary.coords)
+ except:
+ g = np.array(geom.boundary.geoms[0].coords)
+ B.append(g)
+
+
+ B_ix = tuple(itertools.chain.from_iterable(
+ [itertools.repeat(i, x) for i, x in enumerate(list(map(len, B)))]))
+ B = np.concatenate(B)
+ ckd_tree = cKDTree(B)
+
+ dist, idx = ckd_tree.query(A, k=1)
+ idx = itemgetter(*idx)(B_ix)
+
+ gdf = pd.concat(
+ [gdfA, gdfB.loc[idx, gdfB_cols].reset_index(drop=True),
+ pd.Series(dist, name='dist')], axis=1)
+
+ gdf.loc[gdf['dist']>=distance, 'subregion'] = 'Unknown'
+
+ gdf = gdf.drop(columns=['dist'])
+ return gdf
+
+
+if __name__ == "__main__":
+
+ # Define file attributes
+ infile = '/home/pho/Desktop/python_workspace/GrIML/src/griml/test/test_merge_1.shp'
+ regions = '/home/pho/python_workspace/GrIML/other/datasets/drainage_basins/greenland_basins_polarstereo.shp'
+ regions=gpd.read_file(regions)
+
+ regions_dissolve = regions.dissolve(by='subregion')
+
+
+ # Perform conversion
+ a = assign_regions(infile, regions)
+
diff --git a/src/griml/metadata/metadata.py b/src/griml/metadata/metadata.py
deleted file mode 100644
index 9b0ca30..0000000
--- a/src/griml/metadata/metadata.py
+++ /dev/null
@@ -1,38 +0,0 @@
-#!/usr/bin/env python3
-# -*- coding: utf-8 -*-
-
-from griml.metadata import assign_id, assign_sources, assign_certainty, \
- assign_names
-import geopandas as gpd
-
-def add_metadata(iml, names, outfile):
-
- print('Assigning ID...')
- iml = assign_id(iml)
-
- print('Assigning sources...')
- iml = assign_sources(iml)
-
- print('Assigning certainty scores...')
- n = ['S1','S2','ARCTICDEM']
- scores = [0.298, 0.398, 0.304]
- iml = assign_certainty(iml, n, scores)
-
- print('Assigning placenames...')
- iml = assign_names(iml, names)
-
- print('Saving file...')
- iml.to_file(outfile)
- print('Saved to '+str(outfile)+"_metadata.shp")
-
-
-if __name__ == "__main__":
- # indir = "/home/pho/python_workspace/GrIML/other/iml_2017/merged_vectors/griml_2017_inventory.shp"
- indir = "/home/pho/python_workspace/GrIML/other/iml_2017/inspected_vectors/lakes_all-0000000000-0000037888_filtered.shp"
- iml = gpd.read_file(indir)
-
- infile_names = '/home/pho/python_workspace/GrIML/other/datasets/placenames/oqaasileriffik_placenames.shp'
- names = gpd.read_file(infile_names)
-
- outfile = "/home/pho/python_workspace/GrIML/other/iml_2017/metadata_vectors/griml_2017_inventory_final.shp"
- add_metadata(iml, names, outfile)
diff --git a/src/griml/test/greenland_basins_polarstereo.cpg b/src/griml/test/greenland_basins_polarstereo.cpg
new file mode 100644
index 0000000..cd89cb9
--- /dev/null
+++ b/src/griml/test/greenland_basins_polarstereo.cpg
@@ -0,0 +1 @@
+ISO-8859-1
\ No newline at end of file
diff --git a/src/griml/test/greenland_basins_polarstereo.dbf b/src/griml/test/greenland_basins_polarstereo.dbf
new file mode 100644
index 0000000..9d97cd3
Binary files /dev/null and b/src/griml/test/greenland_basins_polarstereo.dbf differ
diff --git a/src/griml/test/greenland_basins_polarstereo.prj b/src/griml/test/greenland_basins_polarstereo.prj
new file mode 100644
index 0000000..2e49174
--- /dev/null
+++ b/src/griml/test/greenland_basins_polarstereo.prj
@@ -0,0 +1 @@
+PROJCS["WGS_1984_NSIDC_Sea_Ice_Polar_Stereographic_North",GEOGCS["GCS_WGS_1984",DATUM["D_WGS_1984",SPHEROID["WGS_1984",6378137.0,298.257223563]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Stereographic_North_Pole"],PARAMETER["False_Easting",0.0],PARAMETER["False_Northing",0.0],PARAMETER["Central_Meridian",-45.0],PARAMETER["Standard_Parallel_1",70.0],UNIT["Meter",1.0]]
\ No newline at end of file
diff --git a/src/griml/test/greenland_basins_polarstereo.shp b/src/griml/test/greenland_basins_polarstereo.shp
new file mode 100644
index 0000000..2773592
Binary files /dev/null and b/src/griml/test/greenland_basins_polarstereo.shp differ
diff --git a/src/griml/test/greenland_basins_polarstereo.shx b/src/griml/test/greenland_basins_polarstereo.shx
new file mode 100644
index 0000000..a63669b
Binary files /dev/null and b/src/griml/test/greenland_basins_polarstereo.shx differ
diff --git a/src/griml/test/process_cli.py b/src/griml/test/process_cli.py
new file mode 100644
index 0000000..915effc
--- /dev/null
+++ b/src/griml/test/process_cli.py
@@ -0,0 +1,132 @@
+#!/usr/bin/env python3
+# -*- coding: utf-8 -*-
+"""
+@author: pho
+"""
+from griml.convert import convert
+from griml.filter import filter_vectors
+from griml.merge import merge_vectors
+from griml.metadata import add_metadata
+from pathlib import Path
+import glob, os
+from argparse import ArgumentParser
+
+def parse_griml_arguments():
+ parser = ArgumentParser(description="Full post-processing workflow for "+
+ "creating ice marginal lake inventory")
+ parser.add_argument('-r', '--root_dir', type=str, required=True,
+ help='Root directory to write files to')
+ parser.add_argument('-i', '--in_dir', type=str, required=True,
+ help='Directory path to input raster files')
+ parser.add_argument('-y', '--year', type=str, required=True,
+ help='Year of inventory')
+ parser.add_argument('-m', '--margin_file', type=str, required=True,
+ help='File path to ice margin for spatial filtering')
+ parser.add_argument('-n', '--names_file', type=str, required=True,
+ help='File path to placenames file for metadata population')
+ parser.add_argument('-p', '--proj', type=str, default='EPSG:3413',
+ required=False, help='Projection (of input and output)')
+ parser.add_argument('-s', '--steps', type=str, default='1111',
+ required=False, help='Define which steps to include in'+
+ ' processing, where each value indicates: convert, '+
+ 'filter, merge, and metadata. If set to zero, the '+
+ 'step associated with that position is skipped')
+
+ args = parser.parse_args()
+ return args
+
+def get_step_flags(a):
+ '''Return step flags'''
+ return a[0],a[1],a[2],a[3]
+
+
+def check_dir(d):
+ '''Check if directory exists and create it if it does not'''
+ if not os.path.exists(d):
+ os.mkdir(d)
+
+
+def griml_process():
+ '''Perform processing workflow'''
+ args = parse_griml_arguments()
+
+ s1, s2, s3, s4 = get_step_flags(args.steps)
+
+ print('Commencing post processing for inventory year ' + args.year)
+ print('Adopted projection: ' + args.proj)
+ print('Writing outputs to ' + args.root_dir)
+
+ root_dir = Path(args.root_dir)
+
+ # Convert to vectors
+ if s1:
+ print('Converting rasters to vectors...')
+
+ src=args.in_dir
+ dest = str(root_dir.joinpath('vectors'))
+ check_dir(dest)
+
+ print('Reading from ' + src)
+ print('Writing to ' + dest)
+
+ band_info = [{'b_number':1, 'method':'VIS', 'source':'S2'},
+ {'b_number':2, 'method':'SAR', 'source':'S1'},
+ {'b_number':3, 'method':'DEM', 'source':'ARCTICDEM'}]
+ start=args.year+'0701'
+ end=args.year+'0831'
+
+ infiles = list(glob.glob(src+'/*.tif'))
+
+ convert(infiles, args.proj, band_info, start, end, str(dest))
+
+
+ # Filter vectors by area and proximity to margin
+ if s2:
+ print('Filtering vectors...')
+
+ src = str(root_dir.joinpath('vectors'))
+ dest = str(root_dir.joinpath('filtered'))
+ check_dir(dest)
+
+ print('Reading from ' + src)
+ print('Writing to ' + dest)
+
+ # margin_buff = gpd.read_file(infile_margin)
+ # margin_buff = margin.buffer(500)
+ # margin_buff = gpd.GeoDataFrame(geometry=margin_buff, crs=margin.crs)
+
+ infiles = list(glob.glob(src+'/*.shp'))
+
+ filter_vectors(infiles, args.margin_file, dest)
+
+
+ # Merge vectors
+ if s3:
+ print('Merging vectors...')
+
+ src = str(root_dir.joinpath('filtered'))
+ dest = root_dir.joinpath('merged/'+args.year+'_merged.shp')
+ check_dir(dest.parent)
+
+ print('Reading from ' + src)
+ print('Writing to ' + str(dest))
+
+ infiles = list(glob.glob(src+'/*.shp'))
+
+ merge_vectors(infiles, str(dest))
+
+
+ # Add metadata
+ if s4:
+ print('Adding metadata...')
+
+ src = str(root_dir.joinpath('merged/'+args.year+'_merged.shp'))
+ dest = root_dir.joinpath('metadata/'+args.year+'_metadata.shp')
+ check_dir(dest.parent)
+
+ print('Reading from ' + src)
+ print('Writing to ' + str(dest))
+
+ add_metadata(src, args.names_file, str(dest))
+
+ print('Finished')
diff --git a/src/griml/test/process_example.py b/src/griml/test/process_example.py
new file mode 100644
index 0000000..c96a4d9
--- /dev/null
+++ b/src/griml/test/process_example.py
@@ -0,0 +1,85 @@
+#!/usr/bin/env python3
+# -*- coding: utf-8 -*-
+"""
+@author: pho
+"""
+from griml.convert import convert
+from griml.filter import filter_vectors
+from griml.merge import merge_vectors
+from griml.metadata import add_metadata
+from pathlib import Path
+import glob
+
+root_dir = Path('/home/pho/python_workspace/GrIML/other/')
+year='2016'
+
+print('Commencing post processing for inventory year ' + year)
+
+# Convert to vectors
+print('Converting rasters to vectors...')
+
+src = str(root_dir.joinpath('iml_2016-2023/rasters/'+year+'_iml'))
+dest = str(root_dir.joinpath('iml_2016-2023/vectors/'+year))
+
+print('Reading from ' + src)
+print('Writing to ' + dest)
+
+proj = 'EPSG:3413'
+band_info = [{'b_number':1, 'method':'VIS', 'source':'S2'},
+ {'b_number':2, 'method':'SAR', 'source':'S1'},
+ {'b_number':3, 'method':'DEM', 'source':'ARCTICDEM'}]
+start=year+'0701'
+end=year+'0831'
+
+infiles = list(glob.glob(src+'/*.tif'))
+
+convert(infiles, proj, band_info, start, end, str(dest))
+
+
+# Filter vectors by area and proximity to margin
+print('Filtering vectors...')
+
+src = dest
+dest = str(root_dir.joinpath('iml_2016-2023/filtered/'+year))
+infile_margin = str(root_dir.joinpath('datasets/ice_margin/gimp_icemask_line_polstereo_simple_buffer.shp'))
+
+print('Reading from ' + src)
+print('Writing to ' + dest)
+
+# margin_buff = gpd.read_file(infile_margin)
+# margin_buff = margin.buffer(500)
+# margin_buff = gpd.GeoDataFrame(geometry=margin_buff, crs=margin.crs)
+
+infiles = list(glob.glob(src+'/*.shp'))
+
+filter_vectors(infiles, infile_margin, dest)
+
+
+# Merge vectors
+print('Merging vectors...')
+
+src = dest
+dest = str(root_dir.joinpath('iml_2016-2023/merged/'+year+'_merged.shp'))
+
+print('Reading from ' + src)
+print('Writing to ' + dest)
+
+infiles = list(glob.glob(src+'/*.shp'))
+
+merge_vectors(infiles, dest)
+
+
+# Add metadata
+print('Adding metadata...')
+
+src = dest
+dest = str(root_dir.joinpath('iml_2016-2023/metadata/'+year+'_metadata.shp'))
+
+print('Reading from ' + src)
+print('Writing to ' + dest)
+
+infile_names = str(root_dir.joinpath('datasets/placenames/oqaasileriffik_placenames.shp'))
+
+add_metadata(src, infile_names, dest)
+
+print('Finished')
diff --git a/src/griml/test/test.py b/src/griml/test/test.py
index 336fdee..24346c3 100644
--- a/src/griml/test/test.py
+++ b/src/griml/test/test.py
@@ -17,7 +17,6 @@ def test_convert(self):
{'b_number':3, 'method':'DEM', 'source':'ARCTICDEM'}]
start='20170701'
end='20170831'
-
infile = os.path.join(os.path.dirname(griml.__file__),'test/test_north_greenland.tif')
convert([infile], proj, band_info, start, end)
@@ -37,7 +36,8 @@ def test_metadata(self):
'''Test metadata population'''
infile1 = os.path.join(os.path.dirname(griml.__file__),'test/test_merge_2.shp')
infile2 = os.path.join(os.path.dirname(griml.__file__),'test/test_placenames.shp')
- add_metadata(infile1, infile2)
+ infile3 = os.path.join(os.path.dirname(griml.__file__),'test/greenland_basins_polarstereo.shp')
+ add_metadata(infile1, infile2, infile3)
if __name__ == "__main__":
unittest.main()
-
-The intent in GrIML is to build upon this pre-existing workflow with new and innovative solutions to form a unified and automated processing chain, with the option to integrate it into a cloud processing platform for efficient big data analysis.
-
-These developments will alleviate the current challenges associated with data-heavy processing (i.e. multi-sensor integration and data retrieval), and ensure detection accuracy with a merged and collated set of established methodologies. These developments will include:
-
-1. Incorporation of the multi-spectral indices classification, backscatter classification, and sink detection methods into one unified processing chain
-
-2. Integration of image analysis from additional sensors into the processing chain, such as Landsat 4-8
-
-3. Automisation of post-processing filtering to remove detached water bodies and misclassifications, using the already-existing 2017 inventory (How et al., 2021) as a training dataset to automatically match and retain existing lakes
-
-4. Adaptation of the workflow for easy transference between offline and cloud processing platforms
-
-## Links
-
-- ESA project outline and fellow information
-
-- Information about the ESA Living Planet Fellowship
-
-- GrIML project description
-
-- 2017 ice marginal lake inventory Scientific Reports paper and dataset
-
-
diff --git a/src/griml.egg-info/SOURCES.txt b/src/griml.egg-info/SOURCES.txt
deleted file mode 100644
index b38c4c2..0000000
--- a/src/griml.egg-info/SOURCES.txt
+++ /dev/null
@@ -1,15 +0,0 @@
-LICENSE
-README.md
-setup.py
-src/griml/__init__.py
-src/griml/dem.py
-src/griml/lake.py
-src/griml/process.py
-src/griml/retrieve.py
-src/griml/sar.py
-src/griml/vis.py
-src/griml.egg-info/PKG-INFO
-src/griml.egg-info/SOURCES.txt
-src/griml.egg-info/dependency_links.txt
-src/griml.egg-info/requires.txt
-src/griml.egg-info/top_level.txt
\ No newline at end of file
diff --git a/src/griml.egg-info/dependency_links.txt b/src/griml.egg-info/dependency_links.txt
deleted file mode 100644
index 8b13789..0000000
--- a/src/griml.egg-info/dependency_links.txt
+++ /dev/null
@@ -1 +0,0 @@
-
diff --git a/src/griml.egg-info/requires.txt b/src/griml.egg-info/requires.txt
deleted file mode 100644
index fe00572..0000000
--- a/src/griml.egg-info/requires.txt
+++ /dev/null
@@ -1,6 +0,0 @@
-earthengine-api
-geopandas
-numpy
-pandas
-scipy
-Shapely
diff --git a/src/griml.egg-info/top_level.txt b/src/griml.egg-info/top_level.txt
deleted file mode 100644
index ba4ef6c..0000000
--- a/src/griml.egg-info/top_level.txt
+++ /dev/null
@@ -1 +0,0 @@
-griml
diff --git a/src/griml/examples/getInventory_funcs.py b/src/griml/examples/getInventory_funcs.py
deleted file mode 100644
index 0dea76c..0000000
--- a/src/griml/examples/getInventory_funcs.py
+++ /dev/null
@@ -1,406 +0,0 @@
-"""
-GrIML processing module playground (experimental)
-
-@author: Penelope How
-"""
-
-import ee, time, sys
-from time import gmtime, strftime
-import requests, zipfile, io, urllib
-from urllib.error import HTTPError
-
-import numpy as np
-from shapely.ops import split
-from shapely.geometry import Polygon, LineString, MultiPolygon
-import geopandas as gpd
-import multiprocessing as mp
-
-sys.path.append('../')
-from retrieve import getScenes, getScene, getInt, getSmooth, getMosaic, \
- getMean, maskImage, splitBBox, getVectors, extractFeatures, getFeatures, \
- getFeaturesSplit
-from sar import filterSARscenes, classifySARimage
-from vis import filterS2scenes, maskS2clouds, renameS2scenes, \
- resampleS2scenes, classifyVISimage, filterLSscenes, maskL8clouds, \
- renameL8scenes
-from dem import getElevation, getSlope, getSinks
-from lake import compileFeatures, assignID, assignSources, assignNames
-
-start=time.time()
-
-#------------------------------------------------------------------------------
-
-# Set AOI
-
-aoi1 = [-50.94, 67.98] # SW test region
-aoi2 = [-49.66, 67.58]
-
-# aoi1 = [-54.68242187500001,68.8415979398901]
-# aoi2 = [-47.21171875000001,60.62185574504481] # SW region
-
-# aoi1 = [-10.297656250000014,59.03190534154833] # All Greenland
-# aoi2 = [-74.98515625000002,83.9867103173338]
-
-print('Loading bounding box')
-aoi = '/home/pho/Desktop/python_workspace/GrIML/other/datasets/aoi/test_mask_4326.shp'
-aoi = gpd.read_file(aoi)
-aoi_poly = aoi.geometry.iloc[0]
-xmin, ymin, xmax, ymax = aoi_poly.bounds
-
-# Set AOI box splitter parameters
-wh=0.3 # Window height
-ww=0.3 # Window width
-oh = 0.05 # Overlap height
-ow = 0.05 # Overlap width
-
-# Set date range
-date1='2017-07-01'
-date2='2017-08-31'
-
-# Set maximum cloud cover
-cloud=50
-
-# Set mask location
-ice_mask = 'users/pennyruthhow/GIMP_iceMask_edit_buffer7km'
-
-# Set output projection
-proj = 'EPSG:3413' # Polar stereographic
-
-
-#--------------------------- Initialise GEE -------------------------------
-
-ee.Initialize()
-print('EE initialized')
-
-# Set the Area of Interest (AOI) through box coordinates
-box = ee.Geometry.Rectangle([xmin, ymin, xmax, ymax])
-
-# Split box into grid for exporting
-bbox = splitBBox(aoi_poly, wh, ww, oh, ow)
-print('grid completed')
-# # Remove boxes that don't overlap with AOI polygon
-# grid = [Polygon([[g[0],g[1]], [g[0],g[3]], [g[2],g[3]],
-# [g[2],g[1]], [g[0],g[1]]]) for g in grid]
-# bbox = [ee.Geometry.Rectangle(min(g.exterior.coords.xy[0]),
-# min(g.exterior.coords.xy[1]),
-# max(g.exterior.coords.xy[0]),
-# max(g.exterior.coords.xy[1])) for g in grid if g.intersects(aoi_poly)]
-
-
-print(f'Computed {len(bbox)} bounding boxes')
-print(f'Total area covered: {aoi_poly.area} sq km')
-
-
-
-
-# bbox = bbox[100:105]
-
-# # Set the Area of Interest (AOI) through box coordinates
-# box = ee.Geometry.Rectangle([aoi1[0], aoi1[1],
-# aoi2[0], aoi2[1]])
-
-# bbox = [ee.Geometry.Rectangle(g[0], g[1], g[2], g[3]) for g in grid]
-
-# print(f'Computed {len(bbox)} bounding boxes from {aoi1}, {aoi2}')
-# print(f'Total area covered: {box.area().getInfo()/10**6} sq km')
-
-
-#--------------------- Get basic ocean mask ---------------------------------
-
-# Retrieve ice-ocean mask
-img = ee.Image("OSU/GIMP/2000_ICE_OCEAN_MASK")
-ocean_mask = img.select('ocean_mask').eq(0)
-# ice_mask = img.select('ice_mask').eq(0)
-
-# imask0 = ice_mask.reduceToVectors(geometryType='polygon', labelProperty='v', scale=10, bestEffort=True)
-# def _mapBuffer(feature):
-# return feature.buffer(-10000)
-# imask01 = imask0.map(_mapBuffer)
-# imask0 = imask01.reduceToImage(properties=['v'], reducer=ee.Reducer.mean())
-
-# sys.exit(1)
-
-# # Construct vector features
-# def getFeaturesSplit(image, bbox):
-# features=[]
-# for b in range(len(bbox)):
-# print(f'Fetching vectors from bbox {b}...')
-# v = getVectors(image, 10, bbox[b])
-# features.append(extractFeatures(v))
-# features = [val for sublist in features for val in sublist]
-# print(f'{len(features)} vector features identified')
-# return features
-
-# def getFeatures(image, bbox):
-# features=[]
-# v = getVectors(image, 10, bbox)
-# features = extractFeatures(v)
-# print(f'{len(features)} vector features identified')
-# return features
-
-# def getParallelFeatures1(image, bbox):
-# # Parallel process vector generation
-# pool = mp.Pool(mp.cpu_count())
-# v = [pool.apply(getVectors, args=(image, 10, bbox[b])) for b in range(len(bbox))]
-# pool.close()
-
-# # Parallel process feature extraction
-# pool = mp.Pool(mp.cpu_count())
-# features = pool.map(extractFeatures, [row for row in v])
-# pool.close()
-
-# features = [val for sublist in features for val in sublist]
-# print(f'{len(features)} vector features identified')
-# return features
-
-# def getDownload(v):
-# link = v.getDownloadURL('csv')
-# req = urllib.request.Request(url=link)
-# try:
-# handler = urllib.request.urlopen(req)
-# except HTTPError as e:
-# handler = e.read()
-# lines = []
-# for l in handler:
-# lines.append(str(l))
-# features_dem.append(lines)
-
-#--------------------------- DEM processing -------------------------------
-
-# Set collection
-dem_col = 'UMN/PGC/ArcticDEM/V3/2m_mosaic'
-
-# Get image collection
-scenes = getScene(dem_col, box)
-print(f'\nScenes gathered from {dem_col}')
-
-# Get elevation and slope
-elevation = getElevation(scenes)
-slope = getSlope(elevation)
-
-elevation = getSmooth(elevation, 110)
-
-# Mask out ocean pixels
-elevation = maskImage(elevation, ocean_mask)
-
-# Get sinks
-elevation = getInt(elevation)
-sinks = getSinks(elevation, 10, 50)
-
-# Remove speckle with smoothing
-sinks = getSmooth(sinks, 50).rename('dem_sinks')
-sinks = getInt(sinks)
-print(f'{dem_col} scenes classified')
-print('Band names: ' + str(sinks.bandNames().getInfo()))
-
-try:
- features_dem = getFeatures(sinks, 10, box)
-except:
- features_dem = getFeaturesSplit(sinks, 10, bbox)
-
-
-#--------------------------- SAR processing -------------------------------
-
-# Set collection
-sar_col = 'COPERNICUS/S1_GRD'
-
-# Get image collection
-scenes = getScenes(sar_col, date1, date2, box)
-if scenes.size().getInfo() > 0:
- scenes = filterSARscenes(scenes)
- print(f'\nScenes gathered from {sar_col}')
- print('Total number of scenes: ', scenes.size().getInfo())
- print('Number of bands per scene: '
- + str(len(scenes.first().bandNames().getInfo())))
- print('Band names: ' + str(scenes.first().bandNames().getInfo()))
-
- # Get average
- aver = getMosaic(scenes, 'HH')
-
- # Mask out ocean pixels
- aver = maskImage(aver, ocean_mask)
-
- # Classify water from SAR average image
- water_sar = classifySARimage(aver, -20, 'HH', 50)
- print(f'{sar_col} scenes classified')
- print('Band names: ' + str(water_sar.bandNames().getInfo()))
-
- try:
- features_sar = getFeatures(water_sar, 10, box)
- except:
- features_sar = getFeaturesSplit(water_sar, 10, bbox)
-
-else:
- features_sar = None
- print(f'\nNo {sar_col} scenes identified between {date1} and {date2}')
-
-#------------------------- VIS processing (S2) -----------------------------
-
-# Set collection
-vis_col1 = "COPERNICUS/S2"
-
-# Get image collection
-scenes = getScenes(vis_col1, date1, date2, box)
-if scenes.size().getInfo() > 0:
- scenes = filterS2scenes(scenes, cloud)
- print(f'\nScenes gathered from {vis_col1}')
- print('Total number of scenes: ', scenes.size().getInfo())
- print('Number of bands per scene: '
- + str(len(scenes.first().bandNames().getInfo())))
- print('Band names: ' + str(scenes.first().bandNames().getInfo()))
-
- # Mask scenes for clouds
- scenes = maskS2clouds(scenes)
- # ee.Terrain.hillshade(image, azimuth, elevation)
-
- # Get average of spectific bands
- scenes = renameS2scenes(scenes)
- scenes = resampleS2scenes(scenes)
- aver = getMean(scenes, ['blue','green','red','vnir','swir1_1','swir2_1'])
- print('Scenes resampled and mosiacked')
- print('Band names: ' + str(aver.bandNames().getInfo()))
-
- # Classify water from VIS average image, and mask out ocean pixels
- water_s2 = classifyVISimage(aver)
- water_s2 = maskImage(water_s2, ocean_mask)
- print(f'{vis_col1} scenes classified')
- print('Band names: ' + str(water_s2.bandNames().getInfo()))
-
- try:
- features_s2 = getFeatures(water_s2, 10, box)
- except:
- features_s2 = getFeaturesSplit(water_s2, 10, bbox)
-else:
- features_s2 = None
- print(f'\nNo {vis_col1} scenes identified between {date1} and {date2}')
-
-#----------------------- VIS processing (Landsat 8) -----------------------
-
-# Set collection
-vis_col2 = "LANDSAT/LC08/C01/T1_TOA"
-
-# Get image collection
-scenes = getScenes(vis_col2, date1, date2, box)
-if scenes.size().getInfo() > 0:
- scenes = filterLSscenes(scenes, cloud)
- print(f'\nScenes gathered from {vis_col2}')
- print('Total number of scenes: ', scenes.size().getInfo())
- print('Number of bands per scene: '
- + str(len(scenes.first().bandNames().getInfo())))
- print('Band names: ' + str(scenes.first().bandNames().getInfo()))
-
- # Mask scenes for clouds
- scenes = maskL8clouds(scenes)
- # ee.Terrain.hillshade(image, azimuth, elevation)
-
- # Get average of spectific bands
- scenes = renameL8scenes(scenes)
- aver = getMean(scenes, ['blue','green','red','vnir','swir1','swir2'])
- print('Scenes resampled and mosiacked')
- print('Band names: ' + str(aver.bandNames().getInfo()))
-
- # Classify water from VIS average image, and mask out ocean pixels
- ndwi = aver.normalizedDifference(['green_mean', 'vnir_mean']).rename('ndwi')
- mndwi = aver.normalizedDifference(['green_mean','swir1_mean']).rename('mndwi')
- aweish = aver.expression('BLUE + 2.5 * GREEN - 1.5 * (VNIR + SWIR1) - 0.25 * SWIR2',
- {'BLUE' : aver.select('blue_mean'),
- 'GREEN' : aver.select('green_mean'),
- 'SWIR1' : aver.select('swir1_mean'),
- 'VNIR' : aver.select('vnir_mean'),
- 'SWIR2' : aver.select('swir2_mean')}).rename('aweish')
- aweinsh = aver.expression('4.0 * (GREEN - SWIR1) - (0.25 * VNIR + 2.75 * SWIR2)',
- {'GREEN' : aver.select('green_mean'),
- 'SWIR1' : aver.select('swir1_mean'),
- 'VNIR' : aver.select('vnir_mean'),
- 'SWIR2' : aver.select('swir2_mean')}).rename('aweinsh')
- bright = aver.expression('(RED + GREEN + BLUE) / 3',
- {'BLUE' : aver.select('blue_mean'),
- 'GREEN' : aver.select('green_mean'),
- 'RED' : aver.select('red_mean')}).rename('bright')
-
- aver = aver.addBands([ndwi, mndwi, aweish, aweinsh, bright])
- classified = aver.expression("(BRIGHT > 5000) ? 0"
- ": (NDWI > 0.3) ? 1 "
- ": (MNDWI < 0.1) ? 0 "
- ": (AWEISH < 2000) ? 0"
- ": (AWEISH > 5000) ? 0"
- ": (AWEINSH < 4000) ? 0"
- ": (AWEINSH > 6000) ? 0"
- ": 1",
- {'NDWI' : aver.select('ndwi'),
- 'MNDWI' : aver.select('mndwi'),
- 'AWEISH' : aver.select('aweish'),
- 'AWEINSH' : aver.select('aweinsh'),
- 'BRIGHT' : aver.select('bright')}).rename('water')
- water_ls8 = classified.updateMask(classified)
- water_ls8 = maskImage(water_ls8, ocean_mask)
- print(f'{vis_col2} scenes classified')
- print('Band names: ' + str(water_ls8.bandNames().getInfo()))
-
- try:
- features_ls8 = getFeatures(water_ls8, 10, box)
- except:
- features_ls8 = getFeaturesSplit(water_ls8, 10, bbox)
-
-else:
- features_ls8 = None
- print(f'\nNo {vis_col2} scenes identified between {date1} and {date2}')
-
-
-#------------------------ Compile geodatabase -----------------------------
-
-iml = compileFeatures([features_s2, features_ls8, features_sar, features_dem],
- ['VIS','VIS','SAR','DEM'],
- [vis_col1, vis_col2, sar_col, dem_col],
- [date1, date2])
-print(f'\nCompiled {iml.shape[0]} geodatabase features')
-
-# Reproject geometries
-iml = iml.to_crs(proj)
-print(f'{iml.shape[0]} unfiltered features')
-
-iml.to_file(f"out/iiml_{date1}_{date2}_{aoi1[0]}_{aoi1[1]}_unfiltered.shp")
-
-
-#-------------------- Filter database by ice margin -----------------------
-
-# Load margin and add buffer
-margin = gpd.read_file("../../../other/datasets/ice_margin/gimp_icemask_line_polstereo.shp")
-margin_buff = margin.buffer(500)
-margin_buff = gpd.GeoDataFrame(geometry=margin_buff, crs=margin.crs)
-
-# Perform spatial join
-iml = gpd.sjoin(iml, margin_buff, how='left')
-iml = iml[iml['index_right']==0]
-iml = iml.drop(columns='index_right')
-print(f'{iml.shape[0]} features within 500 m of margin')
-
-# Filter lakes by area
-iml['area_sqkm'] = iml['geometry'].area/10**6
-iml['length_km'] = iml['geometry'].length/1000
-iml = iml[(iml.area_sqkm >= 0.05)]
-
-# Calculate geometry info
-iml.reset_index(inplace=True, drop=True)
-print(f'{iml.shape[0]} features over 0.05 sq km')
-iml.to_file(f"out/iiml_{date1}_{date2}_{aoi1[0]}_{aoi1[1]}_filtered.shp")
-
-#------------------------- Populate metadata ------------------------------
-
-iml = assignID(iml)
-iml = assignSources(iml)
-
-names = gpd.read_file('../../../other/datasets/placenames/oqaasileriffik_placenames.shp')
-iml = assignNames(iml, names)
-
-#----------------------- Write data to shapefile --------------------------
-
-iml.to_file(f"/home/pho/Desktop/python_workspace/GrIML/other/out/iiml_{date1}_{date2}_{aoi1[0]}_{aoi1[1]}_final.shp")
-print(f'Saved to out/iiml_{date1}_{date2}_{aoi1[0]}_{aoi1[1]}_final.shp')
-
-# Plot lakes
-iml.plot(figsize=(10, 10), alpha=0.5, edgecolor='k')
-
-print('\nFinished')
-end=time.time()
-print('Total run time: ' + strftime("%H:%M:%S", gmtime(round(end-start))))
diff --git a/src/griml/examples/getInventory_obj.py b/src/griml/examples/getInventory_obj.py
deleted file mode 100644
index f8b507c..0000000
--- a/src/griml/examples/getInventory_obj.py
+++ /dev/null
@@ -1,182 +0,0 @@
-"""
-GrIML processing module playground (experimental)
-
-@author: Penelope How
-"""
-
-import time,sys
-from time import gmtime, strftime
-import geopandas as gpd
-
-try:
- sys.path.append('../')
- from process import gee
- from lake import dissolvePolygons, compileFeatures, assignID, assignSources, assignNames
-except:
- from griml.process import gee
- from griml.lake import dissolvePolygons, compileFeatures, assignID, \
- assignSources, assignNames
-
-start=time.time()
-
-#------------------------------------------------------------------------------
-
-# Set AOI box splitter parameters
-wh=0.3 # Window height
-ww=0.3 # Window width
-oh = 0.05 # Overlap height
-ow = 0.05 # Overlap width
-
-# Set date range
-date1='2017-07-01'
-date2='2017-08-31'
-
-# Set output projection
-proj = 'EPSG:3413' # Polar stereographic
-
-# Set AOI
-# aoi = '/home/pho/python_workspace/GrIML/other/datasets/aoi/AOI_mask_split2.shp'
-aoi = '/home/pho/python_workspace/GrIML/other/datasets/aoi/test_mask_4326.shp'
-aoi = gpd.read_file(aoi)#.to_crs('EPSG:4326')
-
-# Load ice margin with buffer
-print('Preparing ice margin buffer...')
-margin_buff = gpd.read_file("/home/pho/python_workspace/GrIML/other/datasets/ice_margin/gimp_icemask_line_polstereo_simple_buffer.shp")
-# margin_buff = margin.buffer(500)
-# margin_buff = gpd.GeoDataFrame(geometry=margin_buff, crs=margin.crs)
-
-# Load name database
-print('Loading placename database...')
-names = gpd.read_file('/home/pho/python_workspace/GrIML/other/datasets/placenames/oqaasileriffik_placenames.shp')
-
-
-#--------------------------- Initialise GEE -------------------------------
-
-parameters = [#{'collection' : 'UMN/PGC/ArcticDEM/V3/2m_mosaic',
- #'smooth' : 100, 'fill' : 100, 'kernel' : 100, 'speckle' : 50},
- # {'collection' : 'UMN/PGC/ArcticDEM/V3/2m',
- # 'smooth' : 100, 'fill' : 100, 'kernel' : 100, 'speckle' : 50},
- {'collection' : 'COPERNICUS/S1_GRD',
- 'polar' : 'HH', 'threshold' : -20, 'smooth' : 50},
- #{'collection' : 'COPERNICUS/S2', `
- #'cloud' : 20},
- {'collection' : 'LANDSAT/LC08/C01/T1_TOA',
- 'cloud' : 20}]
- # {'collection' : 'LANDSAT/LE07/C02/T1_TOA',
- # 'cloud' : 50}]
-
-
-
-for i in range(len(aoi.index)):
- print(f'\nCommencing processing of {list(aoi["catch"])[i]} catchment area')
- aoi_poly = aoi.geometry.iloc[i]
- xmin, ymin, xmax, ymax = aoi_poly.bounds
-
- print('Clipping margin to catchment area...')
- aoi_poly_proj = gpd.GeoDataFrame(geometry=[aoi_poly], crs=aoi.crs).to_crs(proj)
- margin_clip = gpd.clip(margin_buff, aoi_poly_proj)
-
- print('Conducting classifications...')
- proc = gee([date1, date2], aoi_poly, parameters, [wh, ww, oh, ow], True)
-
- print('Processing...')
- water = proc.processAll()
-
- print('Retrieving...')
- features = proc.retrieveAll(water)
-
- # Filter features
- filtered=[]
- for f in features:
- print('Filtering features...')
-
- # Remove duplicates
- print('Dissolving polygons...')
- gdf = gpd.GeoDataFrame(geometry=f, crs='EPSG:4326')
- gdf = dissolvePolygons(gdf)
-
- # Remove lakes below size threshold
- print('Filtering by size...')
- gdf = gdf.set_crs('EPSG:4326')
- gdf = gdf.to_crs(proj)
- gdf['area_sqkm'] = gdf['geometry'].area/10**6
- gdf = gdf[(gdf.area_sqkm >= 0.05)]
-
- # # Remove lakes outside of margin boundary
- # print('Filtering by bounds...')
- # gdf = gpd.sjoin(gdf, margin_clip, how='left')
- # gdf = gdf[gdf['index_right']==0]
- # gdf = gdf.drop(columns='index_right')
- filtered.append(gdf.geometry)
-
-
-#------------------------ Compile geodatabase -----------------------------
-# lakes=[]
-# for f in features:
-# gdf = gpd.GeoDataFrame(geometry=f, crs='EPSG:4326')
-# gdf = dissolvePolygons(gdf)
-# gdf['area'] = gdf['geometry'].area/10**6
-# gdf = gdf[(gdf.area >= 0.05)]
-
-# gdf = gpd.sjoin(gdf, margin_buff, how='left')
-# gdf = gdf[gdf['index_right']==0]
-# gdf = gdf.drop(columns='index_right')
-# lakes.append(list(gdf['geometry']))
-
- # Compile all features
- iml = compileFeatures(filtered,
- ['DEM', 'SAR', 'VIS'],
- ['UMN/PGC/ArcticDEM/V3/2m_mosaic', 'COPERNICUS/S1_GRD',
- 'LANDSAT/LC08/C01/T1_TOA'],
- [date1, date2], proj)
- print(f'\nCompiled {iml.shape[0]} geodatabase features')
-
- # Calculate geometry attributes
- iml['area_sqkm'] = iml['geometry'].area/10**6
- iml['length_km'] = iml['geometry'].length/1000
- iml = iml[(iml.area_sqkm >= 0.05)]
- iml['catch'] = str(list(aoi["catch"])[i])
-
- # Calculate geometry info
- iml.reset_index(inplace=True, drop=True)
-
- print(f'{iml.shape[0]} filtered features')
-
- iml.to_file(f'out/iiml_{date1}_{date2}_{list(aoi["catch"])[i]}_filtered.shp')
-
-
-# #-------------------- Filter database by ice margin -----------------------
-
-# # Perform spatial join
-# aoi_poly_proj = aoi_poly.to_crs(proj)
-# margin_clip = gpd.clip(margin_buff, aoi_poly_proj)
-
-# iml = gpd.sjoin(iml, margin_clip, how='left')
-# iml = iml[iml['index_right']==0]
-# iml = iml.drop(columns='index_right')
-# print(f'{iml.shape[0]} features within 500 m of margin')
-
-# # Calculate geometry info
-# iml.reset_index(inplace=True, drop=True)
-# print(f'{iml.shape[0]} features over 0.05 sq km')
-# iml.to_file(f'out/iiml_{date1}_{date2}_{list(aoi["catch"])[i]}_filtered.shp')
-
-
-#-------------------- Populate metadata and write to file -----------------
-
-# iml = assignID(iml)
- # iml = assignSources(iml)
-
- # iml = assignNames(iml, names)
-
- # iml.to_file(f'out/iiml_{date1}_{date2}_{list(aoi["catch"])[i]}_final.shp')
- # print(f'Saved to out/iiml_{date1}_{date2}_{list(aoi["catch"])[i]}_final.shp')
-
- # # Plot lakes
- # iml.plot(figsize=(10, 10), alpha=0.5, edgecolor='k')
-
-
-#------------------------------------------------------------------------------
-print('\nFinished')
-end=time.time()
-print('Total run time: ' + strftime("%H:%M:%S", gmtime(round(end-start))))
diff --git a/src/griml/metadata/__init__.py b/src/griml/metadata/__init__.py
index 3d16982..7e1e3d8 100644
--- a/src/griml/metadata/__init__.py
+++ b/src/griml/metadata/__init__.py
@@ -1,5 +1,6 @@
from griml.metadata.assign_certainty import *
from griml.metadata.assign_id import *
from griml.metadata.assign_names import *
+from griml.metadata.assign_regions import *
from griml.metadata.assign_sources import *
from griml.metadata.add_metadata import *
diff --git a/src/griml/metadata/add_metadata.py b/src/griml/metadata/add_metadata.py
index 2f15ee2..50e6d9a 100644
--- a/src/griml/metadata/add_metadata.py
+++ b/src/griml/metadata/add_metadata.py
@@ -1,30 +1,17 @@
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
-from griml.metadata import assign_id, assign_sources, assign_certainty, \
- assign_names
from griml.load import load
+from griml.metadata import assign_id, assign_sources, assign_certainty, \
+ assign_names, assign_regions
import geopandas as gpd
-def add_metadata(iml_file, names_file, outfile=None):
- '''Add metadata to collection of features
-
- Parameters
- ----------
- iml_file : str, geopandas.dataframe.DataFrame
- Filepath or geopandas.dataframe.DataFrame object to assign metadata to
-
- Returns
- -------
- iml: geopandas.dataframe.GeoDataFrame
- Geodataframe with metadata
- '''
-
- # Load files
- iml = load(iml_file)
- names = load(names_file)
+def add_metadata(iml, names, regions, outfile=None):
+
+ iml = load(iml)
+ names = load(names)
+ regions = load(regions)
- # Perform metadata steps
print('Assigning ID...')
iml = assign_id(iml)
@@ -35,24 +22,27 @@ def add_metadata(iml_file, names_file, outfile=None):
n = ['S1','S2','ARCTICDEM']
scores = [0.298, 0.398, 0.304]
iml = assign_certainty(iml, n, scores)
+
+ print('Assigning regions...')
+ iml = assign_regions(iml, regions)
print('Assigning placenames...')
iml = assign_names(iml, names)
- # Save to file if given
- if outfile is not None:
+ if outfile:
print('Saving file...')
iml.to_file(outfile)
- print('Saved to '+str(outfile))
-
- return iml
+ print('Saved to '+str(outfile)+"_metadata.shp")
if __name__ == "__main__":
- infile1 = '../test/test_merge_2.shp'
- infile2 = '../test/test_placenames.shp'
- add_metadata(infile1, infile2)
+ # indir = "/home/pho/python_workspace/GrIML/other/iml_2017/merged_vectors/griml_2017_inventory.shp"
+ indir = "/home/pho/python_workspace/GrIML/other/iml_2017/inspected_vectors/lakes_all-0000000000-0000037888_filtered.shp"
+
+
+ infile_names = '/home/pho/python_workspace/GrIML/other/datasets/placenames/oqaasileriffik_placenames.shp'
+ infile_regions = '/home/pho/python_workspace/GrIML/other/datasets/drainage_basins/greenland_basins_polarstereo.shp'
+
- iml = gpd.read_file(infile1)
- names = gpd.read_file(infile2)
- add_metadata(iml, names)
\ No newline at end of file
+ outfile = "/home/pho/python_workspace/GrIML/other/iml_2017/metadata_vectors/griml_2017_inventory_final.shp"
+ add_metadata(indir, infile_names, infile_regions, outfile)
diff --git a/src/griml/metadata/assign_names.py b/src/griml/metadata/assign_names.py
index 9bd294f..c151a48 100644
--- a/src/griml/metadata/assign_names.py
+++ b/src/griml/metadata/assign_names.py
@@ -1,7 +1,18 @@
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
-def assign_names(gdf, gdf_names, distance=500.0):
+import itertools
+from operator import itemgetter
+
+import geopandas as gpd
+import numpy as np
+import pandas as pd
+
+from scipy.spatial import cKDTree
+from shapely.geometry import Point, LineString, Polygon
+from griml.load import load
+
+def assign_names(gdf, gdf_names):
'''Assign placenames to geodataframe geometries based on names in another
geodataframe point geometries
@@ -19,62 +30,41 @@ def assign_names(gdf, gdf_names, distance=500.0):
gdf : pandas.GeoDataFrame
Vectors with assigned IDs
'''
- placenames = _compile_names(gdf_names)
+
+ # Load geodataframes
+ gdf1 = load(gdf)
+ gdf2 = load(gdf_names)
+
+ # Compile placenames into new dataframe
+ names = _compile_names(gdf2)
+ placenames = gpd.GeoDataFrame({"geometry": list(gdf2['geometry']),
+ "placename": names})
- lakename=[]
- for i,v in gdf.iterrows():
-
- geom = v['geometry']
-
- polynames=[]
- for pt in range(len(placenames)):
- if geom.contains(gdf_names['geometry'][pt]) == True:
- polynames.append(placenames[pt])
-
- if len(polynames)==0:
- for pt in range(len(placenames)):
- if gdf_names['geometry'][pt].distance(geom) < distance:
- polynames.append(placenames[pt])
- lakename.append(polynames)
-
- elif len(polynames)==1:
- lakename.append(polynames)
-
- else:
- out=[]
- for p in polynames:
- out.append(p)
- lakename.append(out)
-
- lakeid = gdf['unique_id'].tolist()
- lakename2 = []
+ # Assign names based on proximity
+ a = _get_nearest_point(gdf1, placenames)
+ return a
+
+
+def _get_nearest_point(gdA, gdB, distance=500.0):
+ '''Return properties of nearest point in Y to geometry in X'''
+ nA = np.array(list(gdA.geometry.centroid.apply(lambda x: (x.x, x.y))))
+ nB = np.array(list(gdB.geometry.apply(lambda x: (x.x, x.y))))
+
+ btree = cKDTree(nB)
+ dist, idx = btree.query(nA, k=1)
+ gdB_nearest = gdB.iloc[idx].drop(columns="geometry").reset_index(drop=True)
+ gdf = pd.concat(
+ [
+ gdA.reset_index(drop=True),
+ gdB_nearest,
+ pd.Series(dist, name='dist')
+ ],
+ axis=1)
- for x in gdf.index:
- indx = _get_indices(lakeid, x)
- findname=[]
- for l in indx:
- if len(lakename[l])!=0:
- findname.append(lakename[l])
-
- for i in range(len(indx)):
- if len(findname)==0:
- lakename2.append('')
-
- else:
- unique = set(findname[0])
- unique = list(unique)
-
- if len(unique)==1:
- lakename2.append(findname[0][0])
-
- else:
- out2 = ', '
- out2 = out2.join(unique)
- lakename2.append(out2)
- gdf['placename'] = lakename2
+ gdf.loc[gdf['dist']>=distance, 'placename'] = 'Unknown'
+ gdf = gdf.drop(columns=['dist'])
return gdf
-
def _get_indices(mylist, value):
'''Get indices for value in list'''
return[i for i, x in enumerate(mylist) if x==value]
@@ -95,3 +85,14 @@ def _compile_names(gdf):
else:
placenames.append(None)
return placenames
+
+
+if __name__ == "__main__":
+
+ # Define file attributes
+ infile = '/home/pho/Desktop/python_workspace/GrIML/src/griml/test/test_merge_1.shp'
+ names = '/home/pho/Desktop/python_workspace/GrIML/other/datasets/placenames/oqaasileriffik_placenames_polarstereo.shp'
+
+
+ # Perform conversion
+ a = assign_names(infile, names)
\ No newline at end of file
diff --git a/src/griml/metadata/assign_region.py b/src/griml/metadata/assign_region.py
deleted file mode 100644
index 9bd294f..0000000
--- a/src/griml/metadata/assign_region.py
+++ /dev/null
@@ -1,97 +0,0 @@
-#!/usr/bin/env python3
-# -*- coding: utf-8 -*-
-
-def assign_names(gdf, gdf_names, distance=500.0):
- '''Assign placenames to geodataframe geometries based on names in another
- geodataframe point geometries
-
- Parameters
- ----------
- gdf : pandas.GeoDataFrame
- Vectors to assign uncertainty to
- gdf_names : pandas.GeoDataFrame
- Vector geodataframe with placenames
- distance : int
- Distance threshold between a given vector and a placename
-
- Returns
- -------
- gdf : pandas.GeoDataFrame
- Vectors with assigned IDs
- '''
- placenames = _compile_names(gdf_names)
-
- lakename=[]
- for i,v in gdf.iterrows():
-
- geom = v['geometry']
-
- polynames=[]
- for pt in range(len(placenames)):
- if geom.contains(gdf_names['geometry'][pt]) == True:
- polynames.append(placenames[pt])
-
- if len(polynames)==0:
- for pt in range(len(placenames)):
- if gdf_names['geometry'][pt].distance(geom) < distance:
- polynames.append(placenames[pt])
- lakename.append(polynames)
-
- elif len(polynames)==1:
- lakename.append(polynames)
-
- else:
- out=[]
- for p in polynames:
- out.append(p)
- lakename.append(out)
-
- lakeid = gdf['unique_id'].tolist()
- lakename2 = []
-
- for x in gdf.index:
- indx = _get_indices(lakeid, x)
- findname=[]
- for l in indx:
- if len(lakename[l])!=0:
- findname.append(lakename[l])
-
- for i in range(len(indx)):
- if len(findname)==0:
- lakename2.append('')
-
- else:
- unique = set(findname[0])
- unique = list(unique)
-
- if len(unique)==1:
- lakename2.append(findname[0][0])
-
- else:
- out2 = ', '
- out2 = out2.join(unique)
- lakename2.append(out2)
- gdf['placename'] = lakename2
- return gdf
-
-
-def _get_indices(mylist, value):
- '''Get indices for value in list'''
- return[i for i, x in enumerate(mylist) if x==value]
-
-
-def _compile_names(gdf):
- '''Get preferred placenames from placename geodatabase'''
- placenames=[]
- for i,v in gdf.iterrows():
- if v['Ny_grønla'] != None:
- placenames.append(v['Ny_grønla'])
- else:
- if v['Dansk'] != None:
- placenames.append(v['Dansk'])
- else:
- if v['Alternativ'] != None:
- placenames.append(v['Alternativ'])
- else:
- placenames.append(None)
- return placenames
diff --git a/src/griml/metadata/assign_regions.py b/src/griml/metadata/assign_regions.py
new file mode 100644
index 0000000..6458989
--- /dev/null
+++ b/src/griml/metadata/assign_regions.py
@@ -0,0 +1,86 @@
+#!/usr/bin/env python3
+# -*- coding: utf-8 -*-
+
+import itertools
+from operator import itemgetter
+
+import geopandas as gpd
+import numpy as np
+import pandas as pd
+
+from griml.load import load
+from scipy.spatial import cKDTree
+from shapely.geometry import Point, LineString, Polygon
+
+def assign_regions(gdf, gdf_regions, region_name='region'):
+ '''Assign region to geodataframe geometries based on regions in another
+ geodataframe object
+
+ Parameters
+ ----------
+ gdf : pandas.GeoDataFrame
+ Vectors to assign region name to
+ gdf_regions : pandas.GeoDataFrame
+ Vector geodataframe with regions
+
+ Returns
+ -------
+ gdf : pandas.GeoDataFrame
+ Vectors with assigned IDs
+ '''
+
+ # Load geodataframes
+ gdf1 = load(gdf)
+ gdf2 = load(gdf_regions)
+
+ g = _get_nearest_polygon(gdf1, gdf2)
+
+ return g
+
+
+def _get_nearest_polygon(gdfA, gdfB, gdfB_cols=['subregion'], distance=100000.0):
+ '''Return given properties of nearest polygon in Y to geometry in X'''
+
+ A = np.array(list(gdfA.geometry.centroid.apply(lambda x: (x.x, x.y))))
+ # B = [np.array(geom.boundary.coords) for geom in gdfB.geometry.to_list()]
+
+ B=[]
+ for geom in gdfB.geometry.to_list():
+ try:
+ g = np.array(geom.boundary.coords)
+ except:
+ g = np.array(geom.boundary.geoms[0].coords)
+ B.append(g)
+
+
+ B_ix = tuple(itertools.chain.from_iterable(
+ [itertools.repeat(i, x) for i, x in enumerate(list(map(len, B)))]))
+ B = np.concatenate(B)
+ ckd_tree = cKDTree(B)
+
+ dist, idx = ckd_tree.query(A, k=1)
+ idx = itemgetter(*idx)(B_ix)
+
+ gdf = pd.concat(
+ [gdfA, gdfB.loc[idx, gdfB_cols].reset_index(drop=True),
+ pd.Series(dist, name='dist')], axis=1)
+
+ gdf.loc[gdf['dist']>=distance, 'subregion'] = 'Unknown'
+
+ gdf = gdf.drop(columns=['dist'])
+ return gdf
+
+
+if __name__ == "__main__":
+
+ # Define file attributes
+ infile = '/home/pho/Desktop/python_workspace/GrIML/src/griml/test/test_merge_1.shp'
+ regions = '/home/pho/python_workspace/GrIML/other/datasets/drainage_basins/greenland_basins_polarstereo.shp'
+ regions=gpd.read_file(regions)
+
+ regions_dissolve = regions.dissolve(by='subregion')
+
+
+ # Perform conversion
+ a = assign_regions(infile, regions)
+
diff --git a/src/griml/metadata/metadata.py b/src/griml/metadata/metadata.py
deleted file mode 100644
index 9b0ca30..0000000
--- a/src/griml/metadata/metadata.py
+++ /dev/null
@@ -1,38 +0,0 @@
-#!/usr/bin/env python3
-# -*- coding: utf-8 -*-
-
-from griml.metadata import assign_id, assign_sources, assign_certainty, \
- assign_names
-import geopandas as gpd
-
-def add_metadata(iml, names, outfile):
-
- print('Assigning ID...')
- iml = assign_id(iml)
-
- print('Assigning sources...')
- iml = assign_sources(iml)
-
- print('Assigning certainty scores...')
- n = ['S1','S2','ARCTICDEM']
- scores = [0.298, 0.398, 0.304]
- iml = assign_certainty(iml, n, scores)
-
- print('Assigning placenames...')
- iml = assign_names(iml, names)
-
- print('Saving file...')
- iml.to_file(outfile)
- print('Saved to '+str(outfile)+"_metadata.shp")
-
-
-if __name__ == "__main__":
- # indir = "/home/pho/python_workspace/GrIML/other/iml_2017/merged_vectors/griml_2017_inventory.shp"
- indir = "/home/pho/python_workspace/GrIML/other/iml_2017/inspected_vectors/lakes_all-0000000000-0000037888_filtered.shp"
- iml = gpd.read_file(indir)
-
- infile_names = '/home/pho/python_workspace/GrIML/other/datasets/placenames/oqaasileriffik_placenames.shp'
- names = gpd.read_file(infile_names)
-
- outfile = "/home/pho/python_workspace/GrIML/other/iml_2017/metadata_vectors/griml_2017_inventory_final.shp"
- add_metadata(iml, names, outfile)
diff --git a/src/griml/test/greenland_basins_polarstereo.cpg b/src/griml/test/greenland_basins_polarstereo.cpg
new file mode 100644
index 0000000..cd89cb9
--- /dev/null
+++ b/src/griml/test/greenland_basins_polarstereo.cpg
@@ -0,0 +1 @@
+ISO-8859-1
\ No newline at end of file
diff --git a/src/griml/test/greenland_basins_polarstereo.dbf b/src/griml/test/greenland_basins_polarstereo.dbf
new file mode 100644
index 0000000..9d97cd3
Binary files /dev/null and b/src/griml/test/greenland_basins_polarstereo.dbf differ
diff --git a/src/griml/test/greenland_basins_polarstereo.prj b/src/griml/test/greenland_basins_polarstereo.prj
new file mode 100644
index 0000000..2e49174
--- /dev/null
+++ b/src/griml/test/greenland_basins_polarstereo.prj
@@ -0,0 +1 @@
+PROJCS["WGS_1984_NSIDC_Sea_Ice_Polar_Stereographic_North",GEOGCS["GCS_WGS_1984",DATUM["D_WGS_1984",SPHEROID["WGS_1984",6378137.0,298.257223563]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Stereographic_North_Pole"],PARAMETER["False_Easting",0.0],PARAMETER["False_Northing",0.0],PARAMETER["Central_Meridian",-45.0],PARAMETER["Standard_Parallel_1",70.0],UNIT["Meter",1.0]]
\ No newline at end of file
diff --git a/src/griml/test/greenland_basins_polarstereo.shp b/src/griml/test/greenland_basins_polarstereo.shp
new file mode 100644
index 0000000..2773592
Binary files /dev/null and b/src/griml/test/greenland_basins_polarstereo.shp differ
diff --git a/src/griml/test/greenland_basins_polarstereo.shx b/src/griml/test/greenland_basins_polarstereo.shx
new file mode 100644
index 0000000..a63669b
Binary files /dev/null and b/src/griml/test/greenland_basins_polarstereo.shx differ
diff --git a/src/griml/test/process_cli.py b/src/griml/test/process_cli.py
new file mode 100644
index 0000000..915effc
--- /dev/null
+++ b/src/griml/test/process_cli.py
@@ -0,0 +1,132 @@
+#!/usr/bin/env python3
+# -*- coding: utf-8 -*-
+"""
+@author: pho
+"""
+from griml.convert import convert
+from griml.filter import filter_vectors
+from griml.merge import merge_vectors
+from griml.metadata import add_metadata
+from pathlib import Path
+import glob, os
+from argparse import ArgumentParser
+
+def parse_griml_arguments():
+ parser = ArgumentParser(description="Full post-processing workflow for "+
+ "creating ice marginal lake inventory")
+ parser.add_argument('-r', '--root_dir', type=str, required=True,
+ help='Root directory to write files to')
+ parser.add_argument('-i', '--in_dir', type=str, required=True,
+ help='Directory path to input raster files')
+ parser.add_argument('-y', '--year', type=str, required=True,
+ help='Year of inventory')
+ parser.add_argument('-m', '--margin_file', type=str, required=True,
+ help='File path to ice margin for spatial filtering')
+ parser.add_argument('-n', '--names_file', type=str, required=True,
+ help='File path to placenames file for metadata population')
+ parser.add_argument('-p', '--proj', type=str, default='EPSG:3413',
+ required=False, help='Projection (of input and output)')
+ parser.add_argument('-s', '--steps', type=str, default='1111',
+ required=False, help='Define which steps to include in'+
+ ' processing, where each value indicates: convert, '+
+ 'filter, merge, and metadata. If set to zero, the '+
+ 'step associated with that position is skipped')
+
+ args = parser.parse_args()
+ return args
+
+def get_step_flags(a):
+ '''Return step flags'''
+ return a[0],a[1],a[2],a[3]
+
+
+def check_dir(d):
+ '''Check if directory exists and create it if it does not'''
+ if not os.path.exists(d):
+ os.mkdir(d)
+
+
+def griml_process():
+ '''Perform processing workflow'''
+ args = parse_griml_arguments()
+
+ s1, s2, s3, s4 = get_step_flags(args.steps)
+
+ print('Commencing post processing for inventory year ' + args.year)
+ print('Adopted projection: ' + args.proj)
+ print('Writing outputs to ' + args.root_dir)
+
+ root_dir = Path(args.root_dir)
+
+ # Convert to vectors
+ if s1:
+ print('Converting rasters to vectors...')
+
+ src=args.in_dir
+ dest = str(root_dir.joinpath('vectors'))
+ check_dir(dest)
+
+ print('Reading from ' + src)
+ print('Writing to ' + dest)
+
+ band_info = [{'b_number':1, 'method':'VIS', 'source':'S2'},
+ {'b_number':2, 'method':'SAR', 'source':'S1'},
+ {'b_number':3, 'method':'DEM', 'source':'ARCTICDEM'}]
+ start=args.year+'0701'
+ end=args.year+'0831'
+
+ infiles = list(glob.glob(src+'/*.tif'))
+
+ convert(infiles, args.proj, band_info, start, end, str(dest))
+
+
+ # Filter vectors by area and proximity to margin
+ if s2:
+ print('Filtering vectors...')
+
+ src = str(root_dir.joinpath('vectors'))
+ dest = str(root_dir.joinpath('filtered'))
+ check_dir(dest)
+
+ print('Reading from ' + src)
+ print('Writing to ' + dest)
+
+ # margin_buff = gpd.read_file(infile_margin)
+ # margin_buff = margin.buffer(500)
+ # margin_buff = gpd.GeoDataFrame(geometry=margin_buff, crs=margin.crs)
+
+ infiles = list(glob.glob(src+'/*.shp'))
+
+ filter_vectors(infiles, args.margin_file, dest)
+
+
+ # Merge vectors
+ if s3:
+ print('Merging vectors...')
+
+ src = str(root_dir.joinpath('filtered'))
+ dest = root_dir.joinpath('merged/'+args.year+'_merged.shp')
+ check_dir(dest.parent)
+
+ print('Reading from ' + src)
+ print('Writing to ' + str(dest))
+
+ infiles = list(glob.glob(src+'/*.shp'))
+
+ merge_vectors(infiles, str(dest))
+
+
+ # Add metadata
+ if s4:
+ print('Adding metadata...')
+
+ src = str(root_dir.joinpath('merged/'+args.year+'_merged.shp'))
+ dest = root_dir.joinpath('metadata/'+args.year+'_metadata.shp')
+ check_dir(dest.parent)
+
+ print('Reading from ' + src)
+ print('Writing to ' + str(dest))
+
+ add_metadata(src, args.names_file, str(dest))
+
+ print('Finished')
diff --git a/src/griml/test/process_example.py b/src/griml/test/process_example.py
new file mode 100644
index 0000000..c96a4d9
--- /dev/null
+++ b/src/griml/test/process_example.py
@@ -0,0 +1,85 @@
+#!/usr/bin/env python3
+# -*- coding: utf-8 -*-
+"""
+@author: pho
+"""
+from griml.convert import convert
+from griml.filter import filter_vectors
+from griml.merge import merge_vectors
+from griml.metadata import add_metadata
+from pathlib import Path
+import glob
+
+root_dir = Path('/home/pho/python_workspace/GrIML/other/')
+year='2016'
+
+print('Commencing post processing for inventory year ' + year)
+
+# Convert to vectors
+print('Converting rasters to vectors...')
+
+src = str(root_dir.joinpath('iml_2016-2023/rasters/'+year+'_iml'))
+dest = str(root_dir.joinpath('iml_2016-2023/vectors/'+year))
+
+print('Reading from ' + src)
+print('Writing to ' + dest)
+
+proj = 'EPSG:3413'
+band_info = [{'b_number':1, 'method':'VIS', 'source':'S2'},
+ {'b_number':2, 'method':'SAR', 'source':'S1'},
+ {'b_number':3, 'method':'DEM', 'source':'ARCTICDEM'}]
+start=year+'0701'
+end=year+'0831'
+
+infiles = list(glob.glob(src+'/*.tif'))
+
+convert(infiles, proj, band_info, start, end, str(dest))
+
+
+# Filter vectors by area and proximity to margin
+print('Filtering vectors...')
+
+src = dest
+dest = str(root_dir.joinpath('iml_2016-2023/filtered/'+year))
+infile_margin = str(root_dir.joinpath('datasets/ice_margin/gimp_icemask_line_polstereo_simple_buffer.shp'))
+
+print('Reading from ' + src)
+print('Writing to ' + dest)
+
+# margin_buff = gpd.read_file(infile_margin)
+# margin_buff = margin.buffer(500)
+# margin_buff = gpd.GeoDataFrame(geometry=margin_buff, crs=margin.crs)
+
+infiles = list(glob.glob(src+'/*.shp'))
+
+filter_vectors(infiles, infile_margin, dest)
+
+
+# Merge vectors
+print('Merging vectors...')
+
+src = dest
+dest = str(root_dir.joinpath('iml_2016-2023/merged/'+year+'_merged.shp'))
+
+print('Reading from ' + src)
+print('Writing to ' + dest)
+
+infiles = list(glob.glob(src+'/*.shp'))
+
+merge_vectors(infiles, dest)
+
+
+# Add metadata
+print('Adding metadata...')
+
+src = dest
+dest = str(root_dir.joinpath('iml_2016-2023/metadata/'+year+'_metadata.shp'))
+
+print('Reading from ' + src)
+print('Writing to ' + dest)
+
+infile_names = str(root_dir.joinpath('datasets/placenames/oqaasileriffik_placenames.shp'))
+
+add_metadata(src, infile_names, dest)
+
+print('Finished')
diff --git a/src/griml/test/test.py b/src/griml/test/test.py
index 336fdee..24346c3 100644
--- a/src/griml/test/test.py
+++ b/src/griml/test/test.py
@@ -17,7 +17,6 @@ def test_convert(self):
{'b_number':3, 'method':'DEM', 'source':'ARCTICDEM'}]
start='20170701'
end='20170831'
-
infile = os.path.join(os.path.dirname(griml.__file__),'test/test_north_greenland.tif')
convert([infile], proj, band_info, start, end)
@@ -37,7 +36,8 @@ def test_metadata(self):
'''Test metadata population'''
infile1 = os.path.join(os.path.dirname(griml.__file__),'test/test_merge_2.shp')
infile2 = os.path.join(os.path.dirname(griml.__file__),'test/test_placenames.shp')
- add_metadata(infile1, infile2)
+ infile3 = os.path.join(os.path.dirname(griml.__file__),'test/greenland_basins_polarstereo.shp')
+ add_metadata(infile1, infile2, infile3)
if __name__ == "__main__":
unittest.main()